install.packages(c("sf", "terra", "geodata", "rnaturalearth", "spdep",
"dplyr", "SpatialEpi", "wbstats", "flexdashboard",
"ggplot2", "viridis", "RColorBrewer", "patchwork", "DT",
"leaflet", "mapview", "leafpop", "leafsync", "rasterVis"))
install.packages("INLA",
repos = "https://inla.r-inla-download.org/R/stable", dep = TRUE)Spatial Data Science with R
Paula Moraga, Ph.D.
Assistant Professor of Statistics
King Abdullah University of Science
and Technology (KAUST), Saudi Arabia


Books
Course overview
We will learn statistical methods, modeling approaches, and visualization techniques to analyze spatial data using R
R packages for retrieval, manipulation and visualization of spatial data
Areal, geostatistical and point pattern data
Bayesian spatial models using INLA and SPDE
Interactive visualizations and dashboards to communicate results
Course materials
Spatial Statistics for Data Science: Theory and Practice with R (2023)
https://www.paulamoraga.com/book-spatial/
Geospatial Health Data: Modeling and Visualization with R-INLA & Shiny (2019)
https://www.paulamoraga.com/book-geospatial/
Geospatial Health Data: Modeling and Visualization (2019) http://www.paulamoraga.com/book-geospatial/


Manipulate and transform point, areal, raster data, create maps with R
Fit and interpret Bayesian spatial, spatio-temporal models with INLA, SPDE
Interactive visualizations, reproducible reports, dashboards and Shiny apps
Spatial Statistics for Data Science: Theory and Practice with R (2023) http://www.paulamoraga.com/book-spatial/
Spatial data: types, retrieval, manipulation and visualization. Statistical methods and models to analyze spatial data using R
Areal data: spatial neighborhood matrices, autocorrelation, models
Geostatistical data: interpolation, kriging, model-based geostatistics
Point patterns: intensity estimation, clustering, point process models
Reproducible examples in environment, ecology, epidemiology, crime, real state

R packages
Course schedule
| 9:30 am - 10:00 am | Introduction geospatial data and methods |
| 10:00 am - 11:00 am | Areal data modeling |
| 11:00 am - 11:15 am | Break |
| 11:15 am - 12:00 pm | Geostatistical data modeling |
| 12:00 pm - 12:30 pm | Interactive dashboards |
Geospatial data and methods

John Snow’s map of cholera, London, 1854
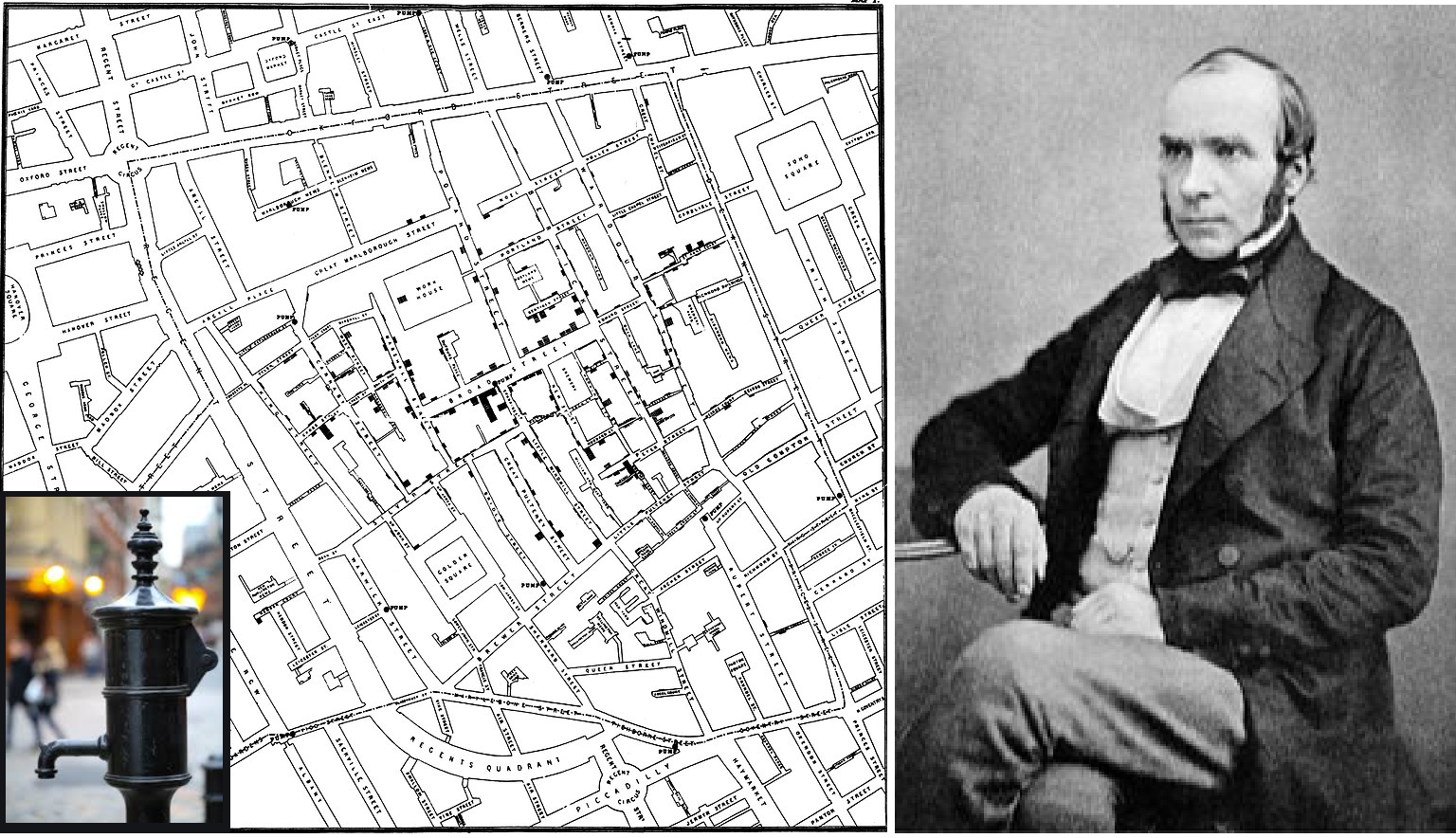
Geospatial methods for disease surveillance
Geospatial methods use data on disease cases, population at risk, and risk factors such as environmental, climate and socio-economic variables to
- Understand geographic and temporal patterns
- Identify potential risk factors
- Highlight high risk areas and detect clusters
- Measure inequalities
- Early detection of outbreaks
🗣 Results can be communicated using maps and other visualizations
✍️ Results guide decision-makers to better allocate limited resources and to design strategies for disease prevention and control
🌍 Many methods useful in fields such as ecology, environment, criminology
Types of spatial data
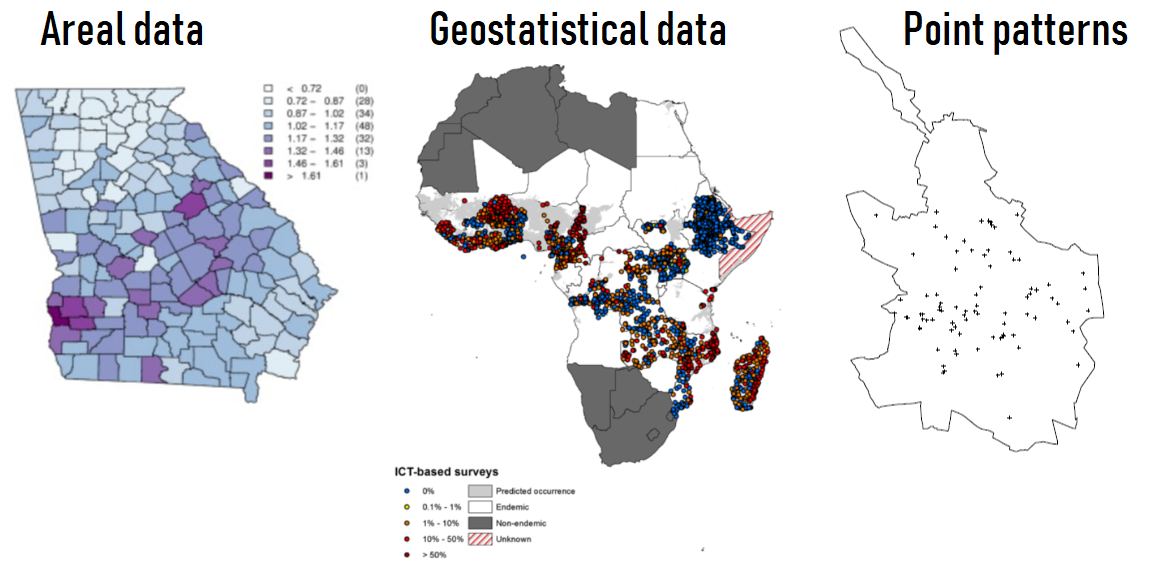
Moraga and Lawson, Computational Statistics & Data Analysis, 2012
Moraga et al., Parasites & Vectors, 2015
Moraga and Montes, Statistics in Medicine, 2011
Areal data
Standardized Mortality Ratio (SMR) is often used to estimate disease risk
\[ SMR_i = \frac{Y_i}{E_i} = \frac{\mbox{number observed cases in area } i}{\mbox{number expected cases in area } i}\]
\(SMR_i = 1\) same number observed as expected
\(SMR_i > 1\) more observed than exp. (high risk)
\(SMR_i < 1\) less observed than exp. (low risk)
Limitations
SMRs easy to calculate but may be misleading and unreliable in areas with small populations or rare diseases. Models enable to incorporate covariates & borrow information from neighboring areas to obtain smoothed relative risks
Areal models
Model to estimate disease relative risk \(\theta_i\) in areas \(i=1,\ldots,n\)
\[Y_i|\theta_i \sim Poisson(E_i \times \theta_i)\] \[\log(\theta_i) = \boldsymbol{z}_i \boldsymbol{\beta} + u_i + v_i\]
- \(Y_i\) observed cases, \(E_i\) expected cases, \(\theta_i\) relative risk in area \(i\)
Fixed effects quantify the effects of the covariates on the disease risk
- \(\boldsymbol{z}_i = (1, z_{i1}, \ldots, z_{ip})\) intercept and cov. \(\beta = (\beta_0, \beta_1, \ldots, \beta_p)'\) coeff.
Random effects represent residual variation not explained by the covariates
- \(u_i\): structured spatial effect to account for spatial dependence between relative risks (close areas show more similar risk than not close areas)
- \(v_i\): unstructured effect to account for independent noise
Spatial neighborhood matrix
Inference using INLA
\[Y_i|\theta_i \sim Poisson(E_i \times \theta_i)\] \[\log(\theta_i) = \boldsymbol{z}_i \boldsymbol{\beta} + u_i + v_i\]
library(INLA)
formula <- Y ~ cov +
f(idareau, model = "besag", graph = g, scale.model = TRUE) +
f(idareav, model = "iid")
res <- inla(formula, family = "poisson", data = map@data,
E = E, control.predictor = list(compute = TRUE))
# Posterior mean and 95% CI
map$RR <- res$summary.fitted.values[, "mean"]
map$LL <- res$summary.fitted.values[, "0.025quant"]
map$UL <- res$summary.fitted.values[, "0.975quant"]Visualization
library(leaflet)
pal <- colorNumeric(palette = "YlOrRd", domain = map$SIR)
leaflet(map) %>% addTiles() %>%
addPolygons(color = "grey", weight = 1, fillColor = ~ pal(SIR)) %>%
addLegend(pal = pal, values = ~SIR, title = "SIR")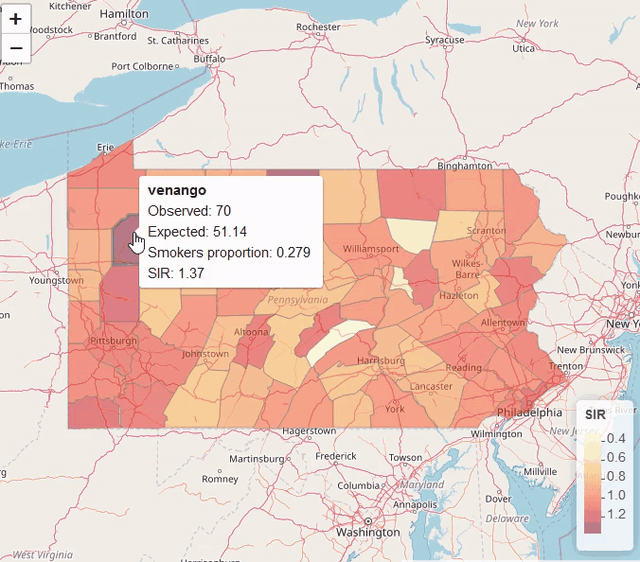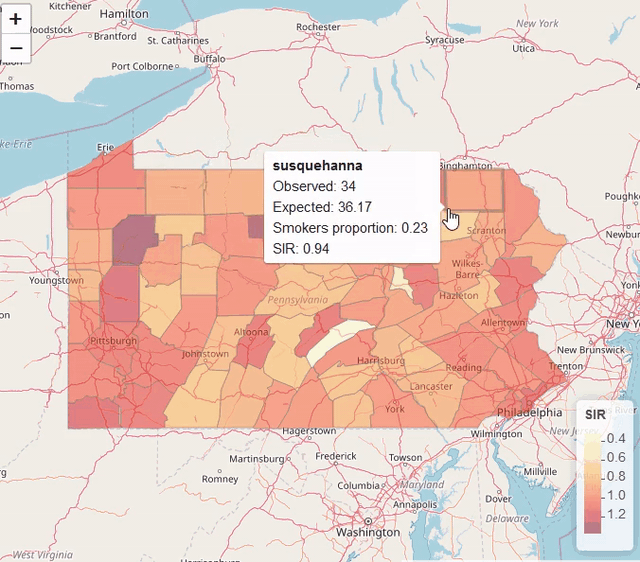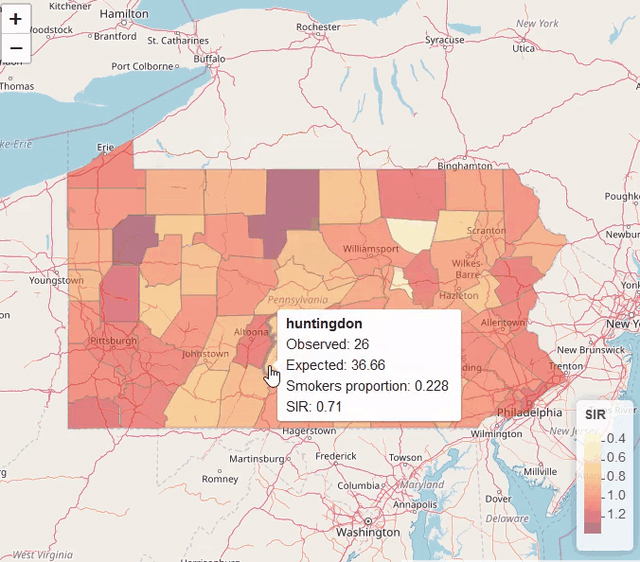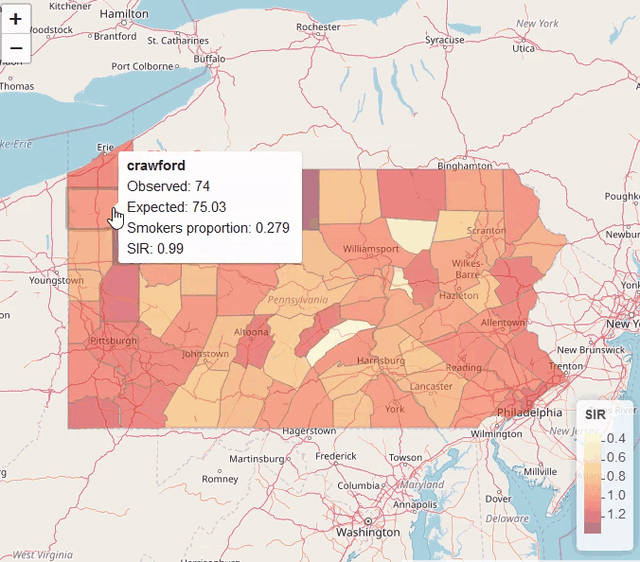
Geostatistical data
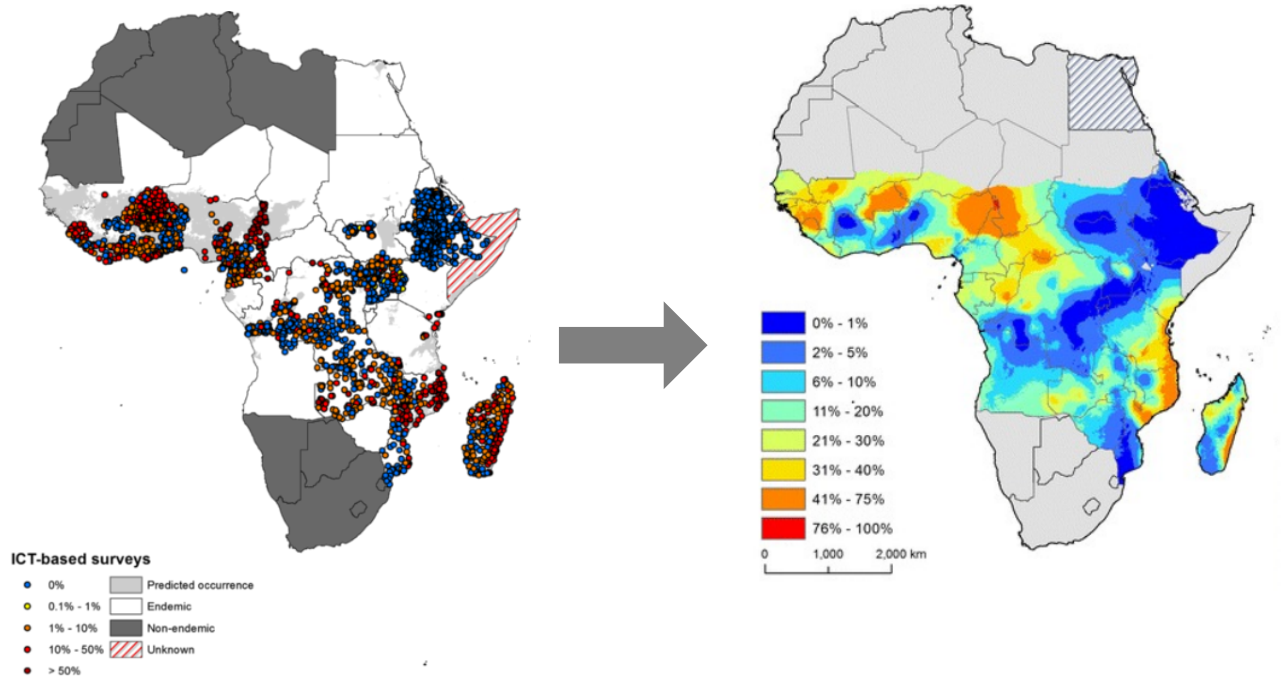
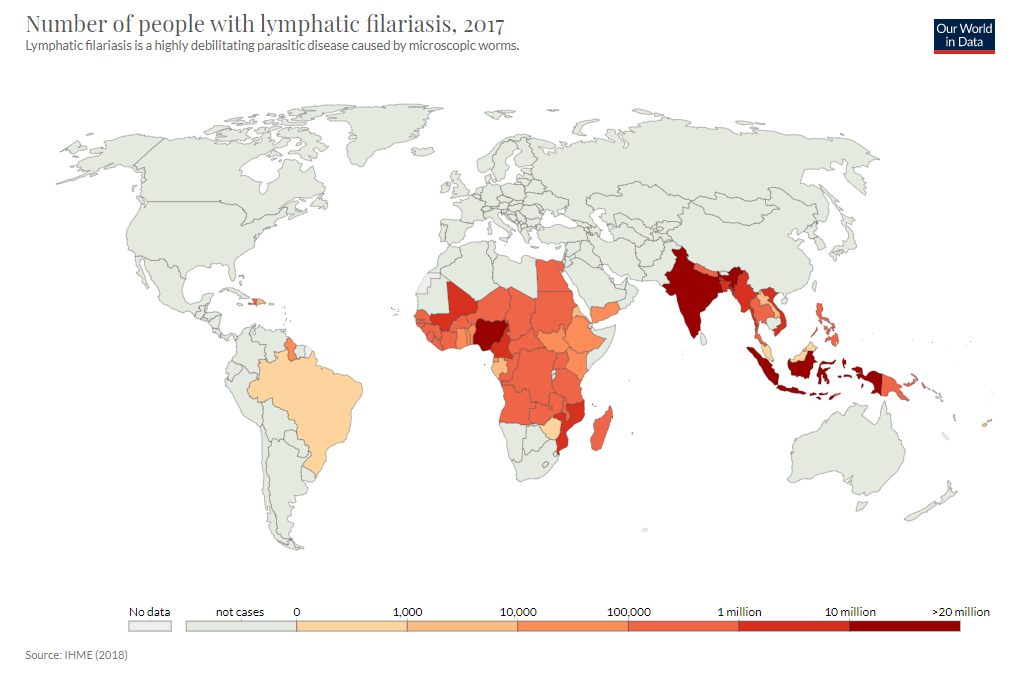
Geospatial modeling of lymphatic filariasis prevalence in sub-Saharan Africa
Lymphatic filariasis caused by microscopic worms and transmitted by mosquitoes
Main strategy against the disease is Mass Drug Administration. Resources are limited and need to decide which areas most in need
Geospatial modeling of lymphatic filariasis

3197 LF prevalence surveys, 1990-2014
Geostatistical models
Models to predict prevalence at unsampled locations
\[Y_i|P(\boldsymbol{x}_i)\sim \mbox{Binomial} (n_i, P(\boldsymbol{x}_i))\] \[\mbox{logit}(P(\boldsymbol{x}_i)) = \boldsymbol{z}_i \boldsymbol{\beta} + S(\boldsymbol{x}_i) + u_i\]
\(Y_i\) number people positive, \(n_i\) number people tested, \(P(\boldsymbol{x}_i)\) prevalence at \(\boldsymbol{x}_i\)
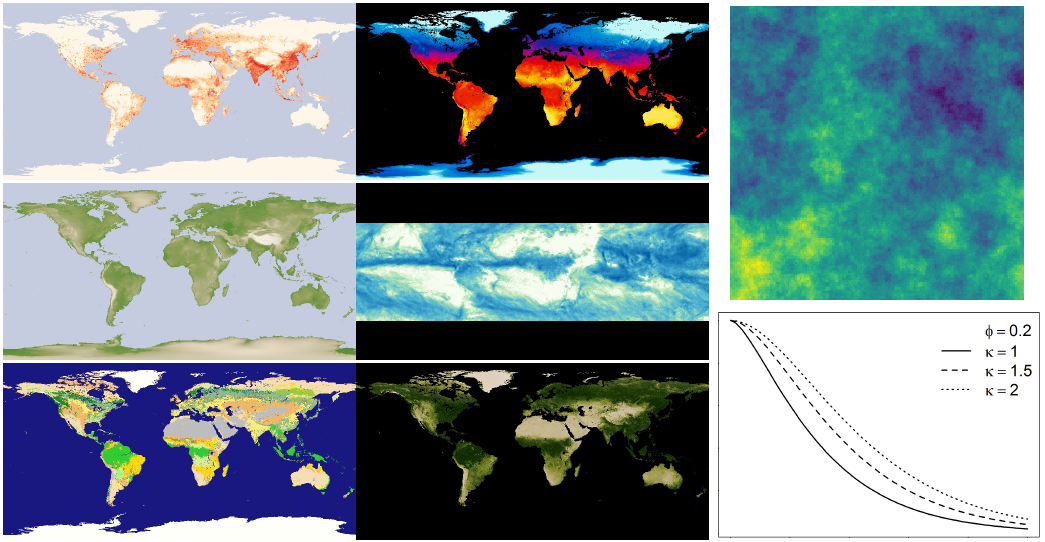
Covariates based on characteristics known to affect disease transmission (temperature, precipitation, vegetation, elevation, land cover, population density, etc.)
Random effects model residual variation not explained by covariates
Triangulated mesh for SPDE approach
library(INLA)
mesh <- inla.mesh.2d(
loc = coo, offset = c(50, 100),
cutoff = 1, max.edge = c(30, 60))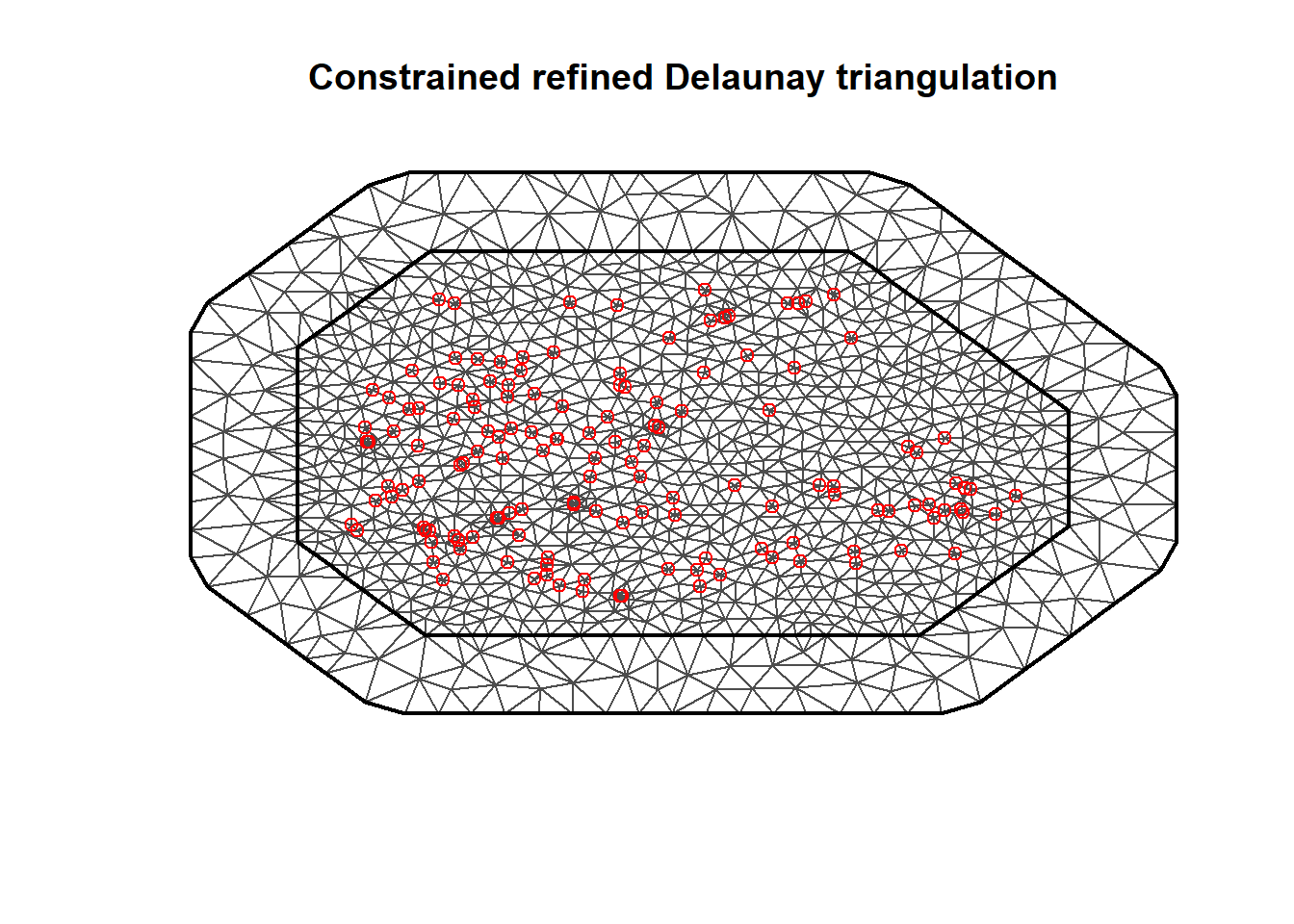
Spatial covariates
Inference using INLA and SPDE
\[Y_i|P(\boldsymbol{x}_i)\sim \mbox{Binomial} (n_i, P(\boldsymbol{x}_i))\] \[\mbox{logit}(P(\boldsymbol{x}_i)) = \boldsymbol{z}_i \boldsymbol{\beta} + S(\boldsymbol{x}_i) + u_i\]
library(INLA)
formula <- y ~ 0 + b0 + alt + f(s, model=spde) + f(idu, model="iid")
res <- inla(formula, family = "binomial", Ntrials = numtrials,
control.family = list(link = "logit"),
data = inla.stack.data(stk.full),
control.predictor = list(compute = TRUE, link = 1,
A = inla.stack.A(stk.full)))
index <- inla.stack.index(stack = stk.full, tag = "pred")$data
# Posterior mean and 95% CI
prev_mean <- res$summary.fitted.values[index, "mean"]
prev_ll <- res$summary.fitted.values[index, "0.025quant"]
prev_ul <- res$summary.fitted.values[index, "0.975quant"]Point patterns
Assume point pattern \(\{s_i: i=1, \ldots, n\}\) has been generated as a realization of a point process \(Z = \{Z(s): s \in \mathbb{R}^2\}\)
A point process model can be used to estimate the intensity of events, identify patterns in the distribution of the observed locations, and learn about the correlation between the locations and spatial covariates
Intensity of the process
Intensity of the process: mean number of events per unit area at location \(s\) \[\lambda(s) = lim_{|ds|\rightarrow 0} \frac{E[N(ds)]}{|ds|}\]
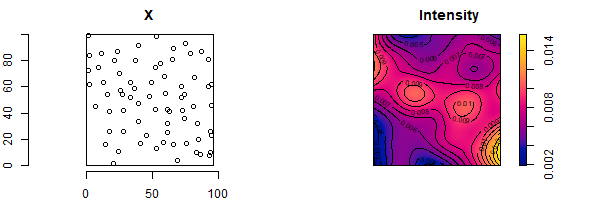
Homogeneous Poisson process
Simple model that assumes events are equally likely to occur at any location within the study area, independent of the locations of other events
Log-Gaussian Cox process
Typically used to model phenomena that are environmentally driven. Poisson process with a varying intensity which is itself a stochastic process of the form \(\Lambda(s) = exp(\eta(s))\), \(\eta = \{\eta(s): s \in \mathbb{R}^2\}\) spatial Gaussian process
number of events in any region \(A\) is distributed as \(Po(\int_A \Lambda(s)ds)\)
locations indep. random sample with probability density proportional to \(\Lambda(s)\)
Types of spatial data

Moraga and Lawson, Computational Statistics & Data Analysis, 2012
Moraga et al., Parasites & Vectors, 2015
Moraga and Montes, Statistics in Medicine, 2011
Exercise
State whether each of the following is an example of areal data, geostatistical data or a point pattern.
Geographical locations of the home addresses of a sample of cardiovascular patients in Italy.
Yesterday’s mid-day temperature at the centres of fifty cities in Germany.
Carbon dioxide levels measured at twenty locations near a road.
The positions of hair follicles on a human head.
Crop yields from each of thirteen adjacent rows of wheat in a field.
Positions of snooker balls on a snooker table after one shot has been played.
The average house price in each postcode region in Paris.
Spatial data and
visualization with R
Spatial data in R
Spatial data can be represented using vector and raster data
Vector data displays points, lines and polygons, and associated information
Examples: locations of monitoring stations, road networks, municipalities
Raster data are regular grids with cells of equal size that are used to store values of spatially continuous phenomena
Examples: elevation, temperature, air pollution values
R packages: sf (vector data) and terra (raster and vector data)
sf to work with vector data
Vector data are often represented using a data format called shapefile.

Shapefile to store vector data
A shapefile is a collection files.

terra to work with raster (and vector) data
Raster data often come in GeoTIFF format which has extension .tif.

R packages to download open spatial data
Chapter 6 Book Spatial Statistics for Data Science
https://rspatialdata.github.io/
| Data | R package | Database |
|---|---|---|
| Administrat. boundaries | rnaturalearth | rnaturalearth |
| Population | wopr | WorldPop |
| OpenStreetMap | osmdata | OpenStreetMap (OSM) |
| Elevation | elevatr | AWS Terrain Tiles |
| Temperature | raster | WorldClim |
| Humidity | nasapower | NASA-POWER Project |
| Vegetation | MODIStsp | MODIS |
| Land cover | MODIStsp | MODIS |
| Malaria | malariaAtlas | Malaria Atlas Project (MAP) |
Coordinate Reference Systems (CRS)
unprojected or geographic: Latitude and Longitude for referencing location on the ellipsoid Earth
(decimal degrees (DD) or degrees, minutes, and seconds (DMS))projected: Easting and Northing for referencing location on 2-dimensional representation of Earth
Common projection: Universal Transverse Mercator (UTM)
Location is given by the zone number (60 zones), hemisphere (north or south), and Easting and Northing coordinates in the zone in meters
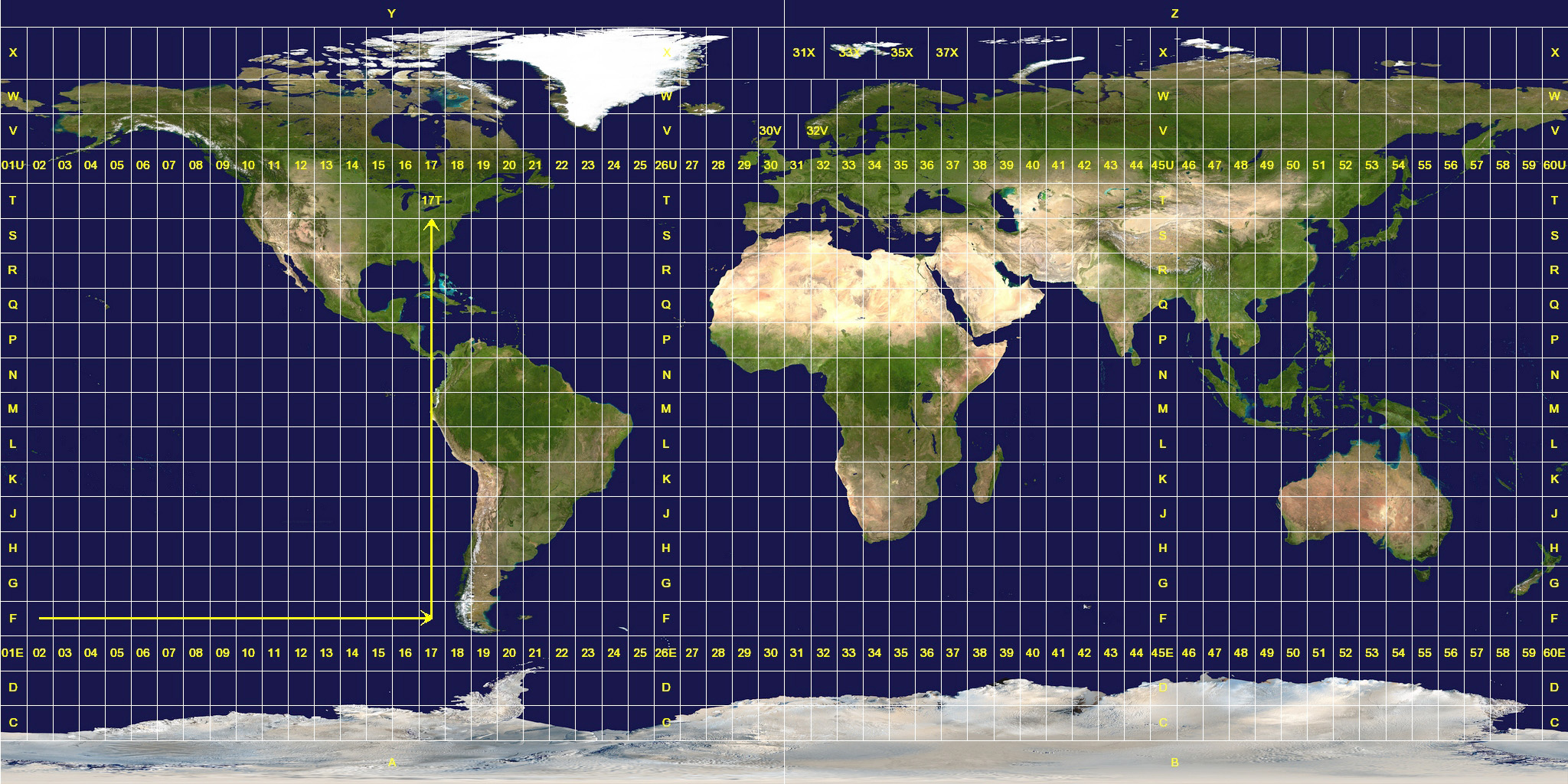
Coordinate Reference Systems (CRS)
Most common CRSs can be specified with their EPSG (European Petroleum Survey Group) codes
EPSG 4326 refers to the geographic CRS (latitude and longitude)
Common spatial projections: https://spatialreference.org/ref/
Geographic coordinates of New York City, USA:
| Latitude | Longitude | |
|---|---|---|
| Degrees, Minutes and Seconds | 40° 43’ 50.1960’’ North | 73° 56’ 6.8712’’ West |
| Decimal degrees (North/South and West/East) | 40.730610° North | 73.935242° West |
| Decimal degrees (Positive/Negative) | 40.730610 | -73.935242 |
Interactive visualizations
and dashboards to communicate results
Visualization
Making maps with R
Chapter 5 Book Spatial Statistics for Data Science
HTML widgets
HTML widgets are interactive web visualizations built with JavaScript
Leaflet
http://rstudio.github.io/leaflet/

Dygraphs
http://rstudio.github.io/dygraphs/

DataTable
Reproducible documents with R Markdown
R Markdown can be used to turn our analysis into fully reproducible documents that can be shared with others. Output formats include HTML, PDF or Word. An R Markdown file is written with Markdown syntax with embedded R code, and can include narrative text, tables and visualizations
http://www.paulamoraga.com/book-geospatial/sec-rmarkdown.html
Interactive dashboards with flexdashboard
flexdashboard uses R Markdown to publish a group of related data visualizations as a dashboard
http://www.paulamoraga.com/book-geospatial/sec-flexdashboard.html
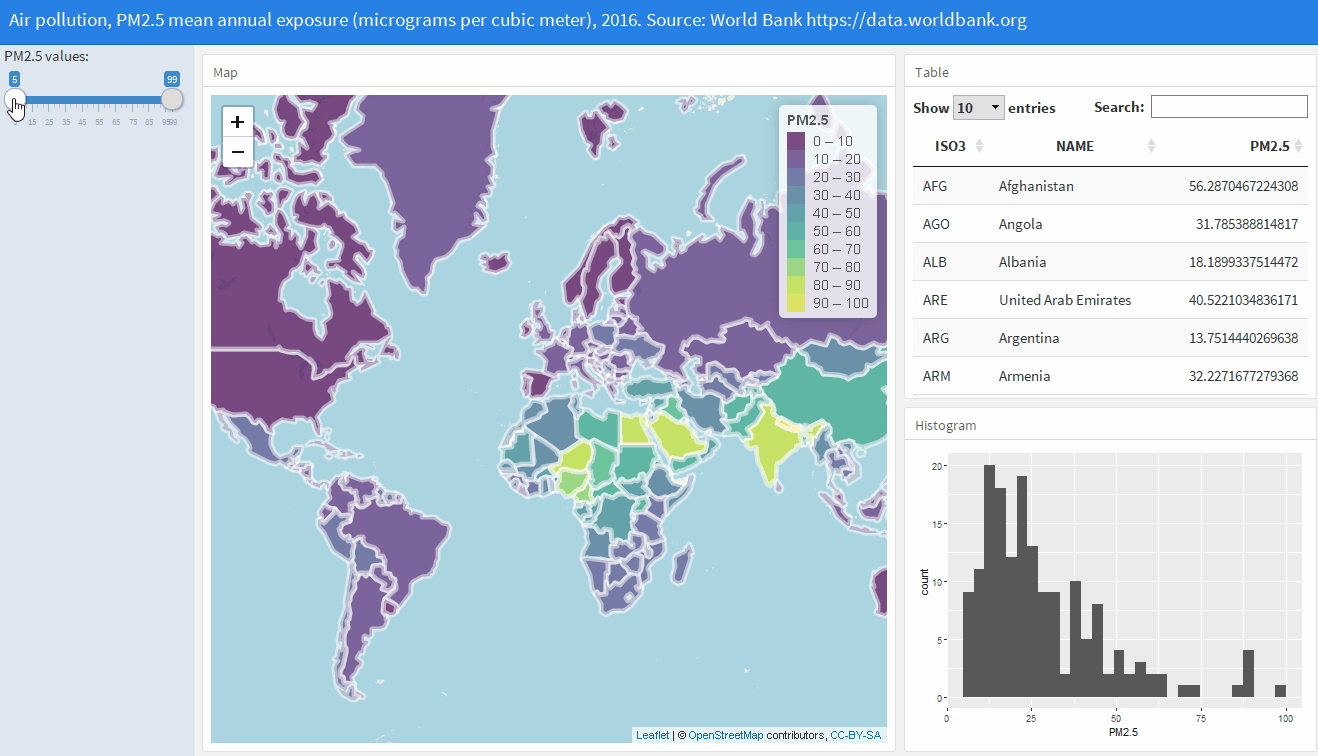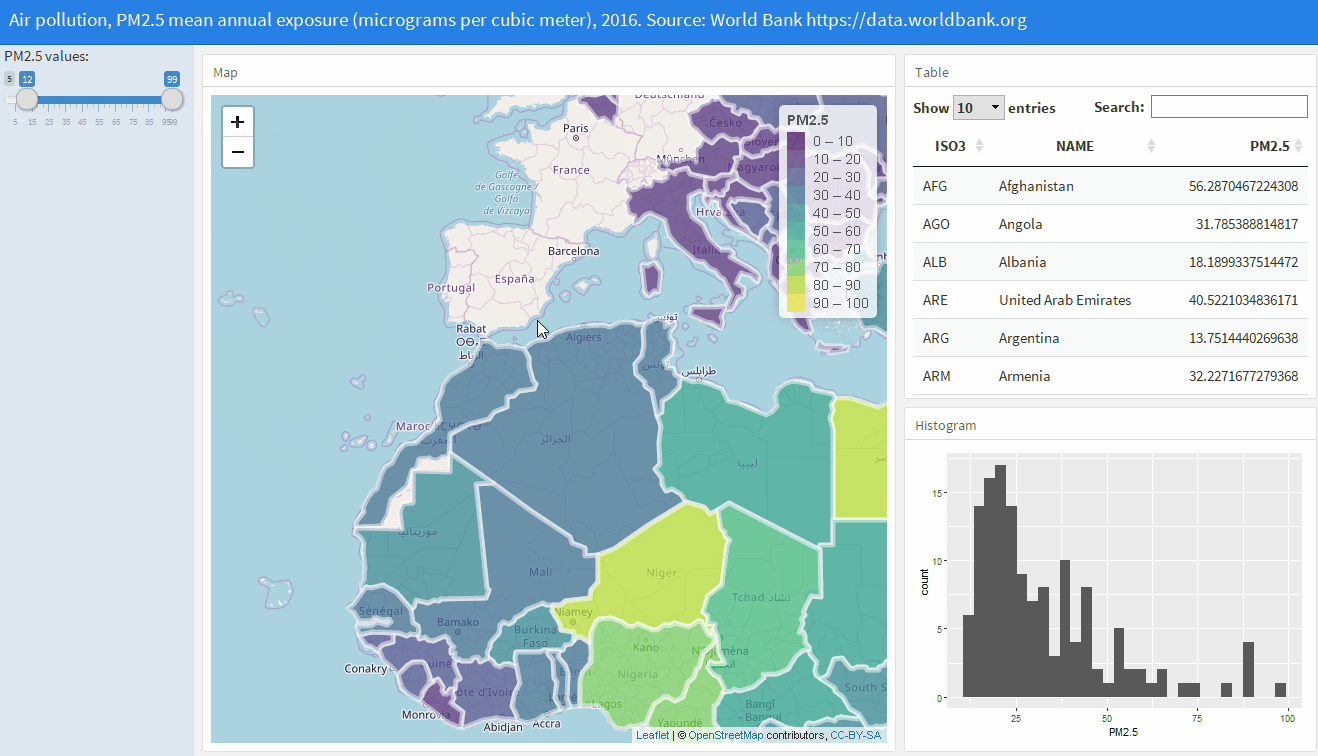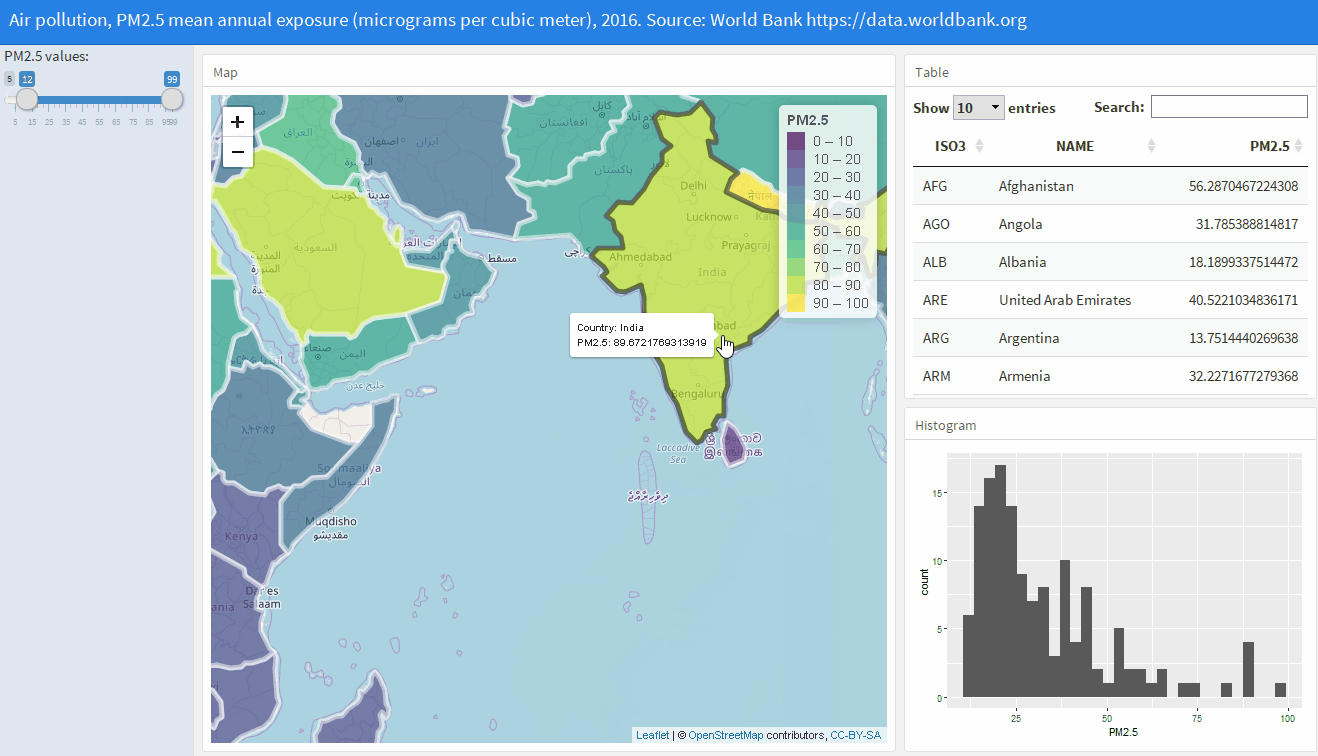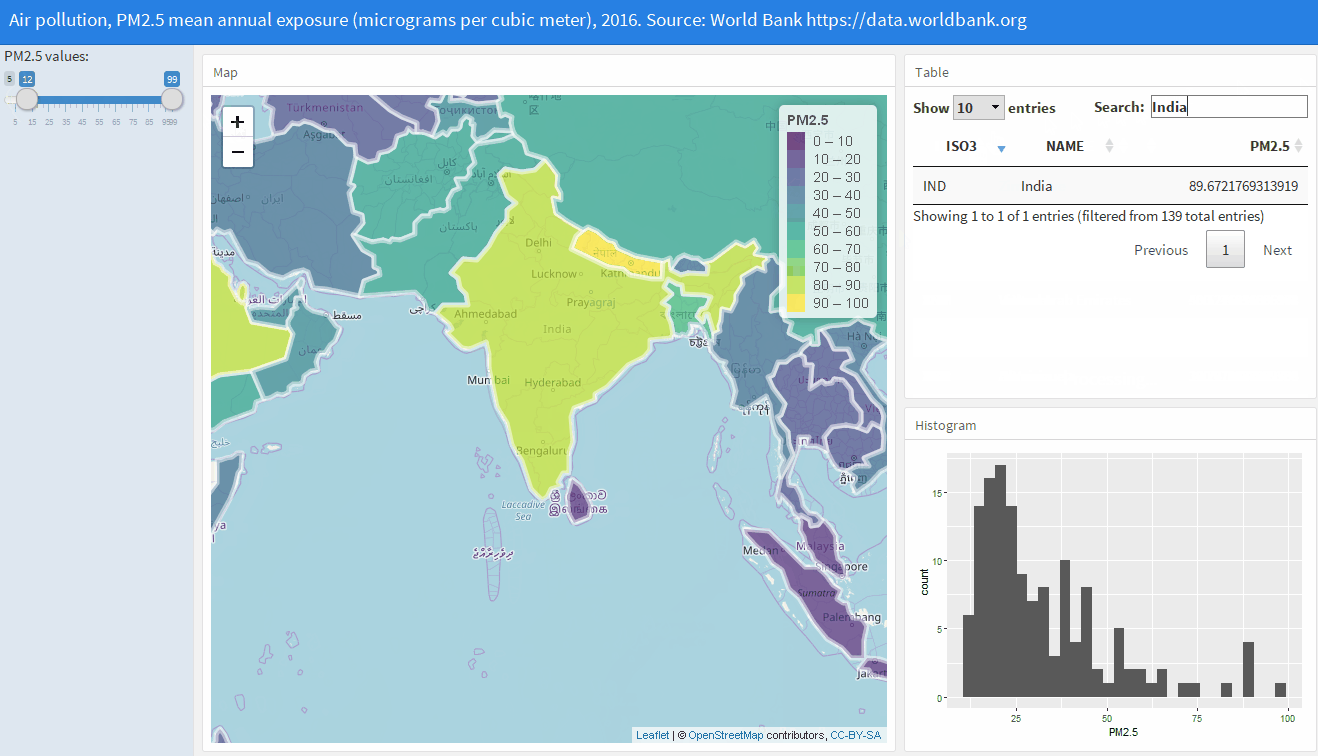
Shiny web applications
Shiny is a web application framework for R that enables to build interactive web applications
http://www.paulamoraga.com/book-geospatial/sec-shiny.html
SpatialEpiApp is a Shiny app for disease risk estimation, cluster detection, and interactive visualization
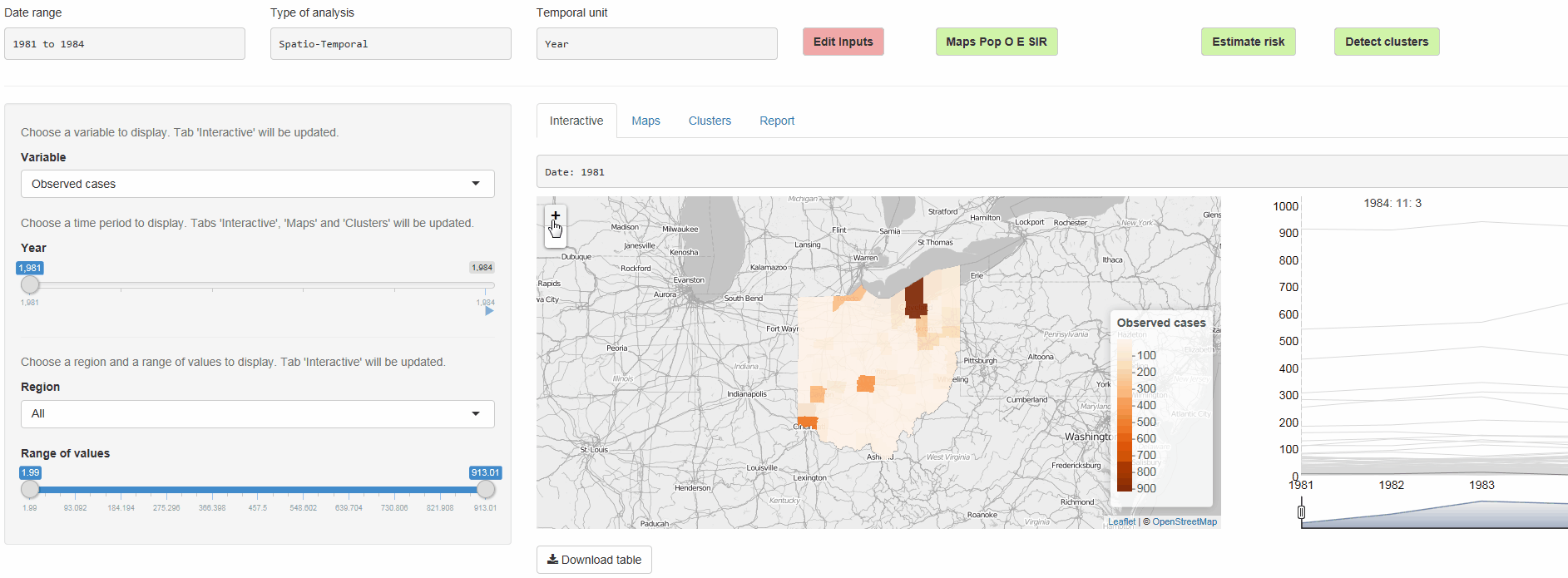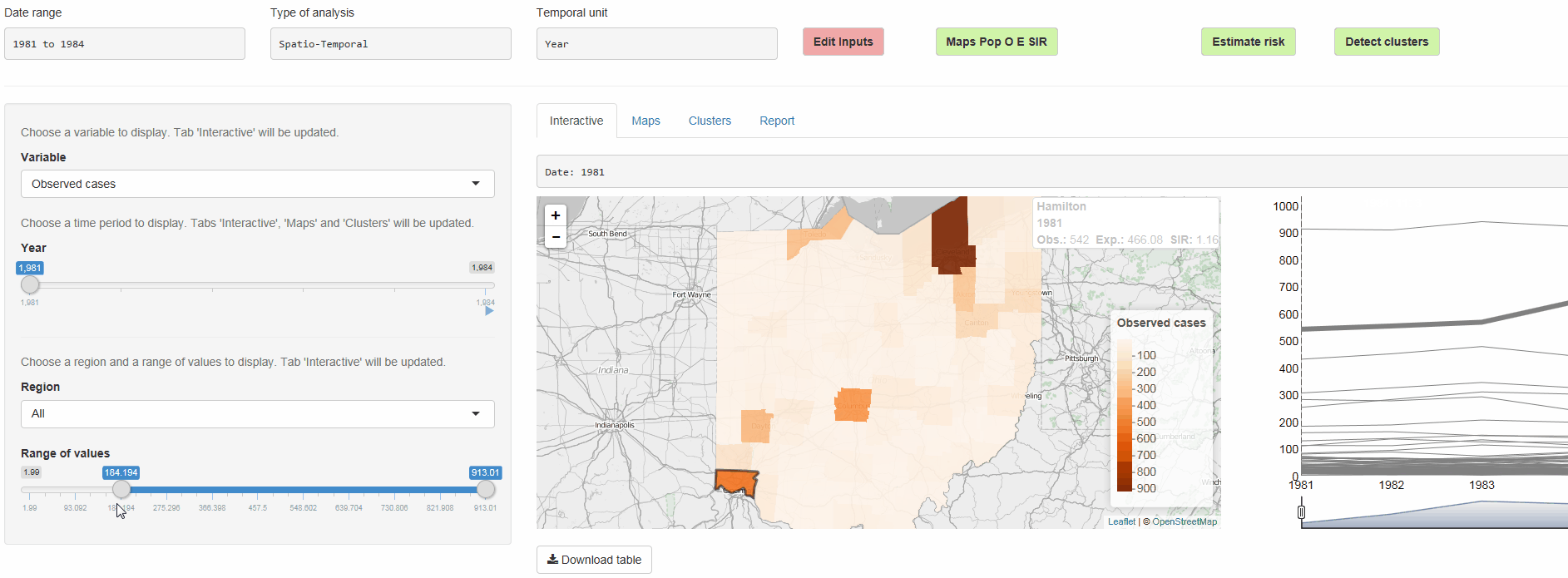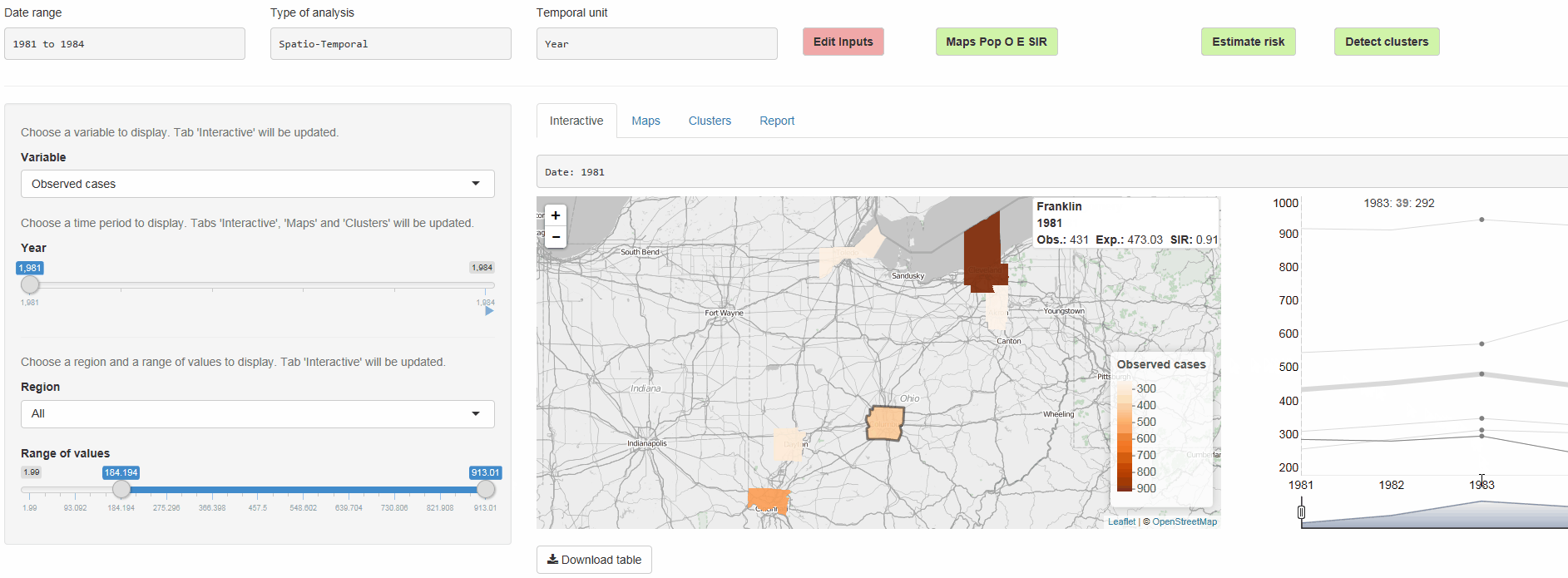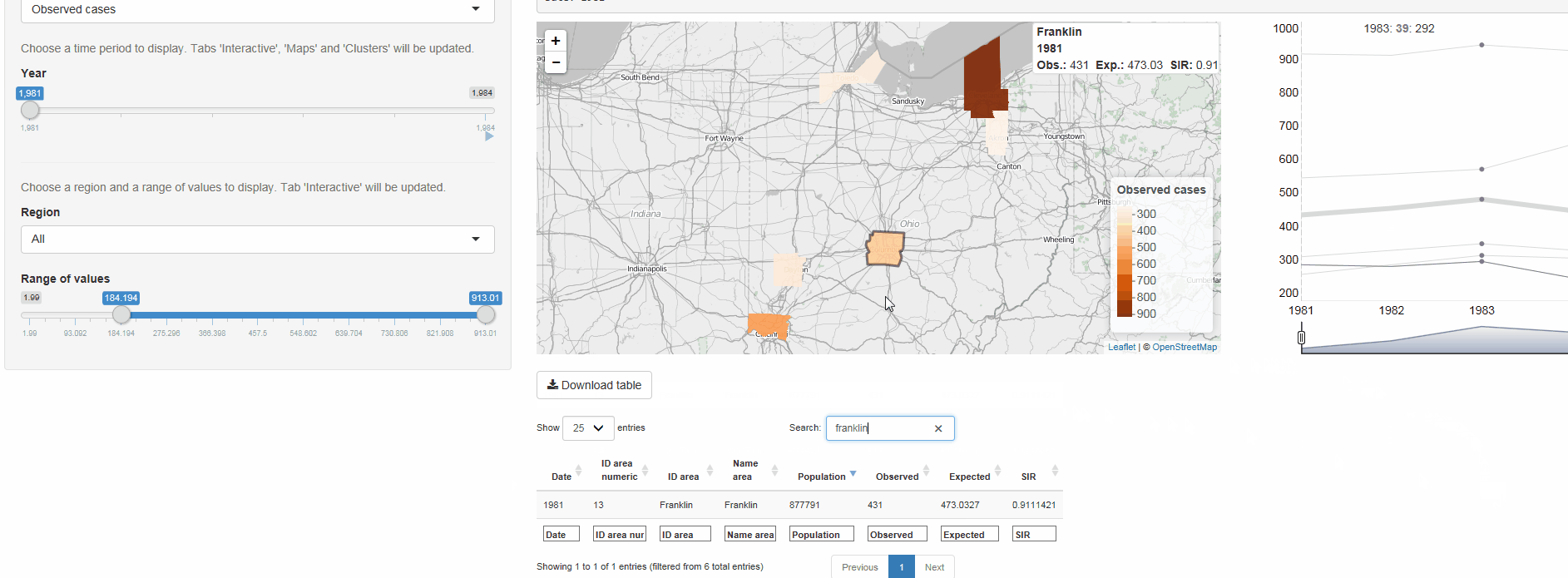
Geospatial modeling
Integrated nested Laplace approximation (INLA)
INLA
Integrated nested Laplace approximation (INLA) is a computational approach to perform approximate Bayesian inference in latent Gaussian models such as generalized linear mixed models and spatial and spatio-temporal models
INLA uses a combination of analytical approximations and numerical integration to obtain approximated posterior distributions of parameters and is very fast compared to MCMC. The combination of INLA and the stochastic partial differential equation (SPDE) approaches permits to analyze point data
INLA and SPDE can be applied with the R package R-INLA
Resources
INLA website http://www.r-inla.org/
Geospatial Health Data: Modeling and Visualization with R-INLA and Shiny. Moraga. CRC Press, 2019 https://www.paulamoraga.com/book-geospatial/
Advanced Spatial Modeling with SPDE Using R and INLA. Krainski et al. CRC Press, 2019 https://becarioprecario.bitbucket.io/spde-gitbook/
Latent Gaussian models
Observations belong to an exponential family with mean \(\mu_i=g^{-1}(\eta_i)\)
(e.g. Gaussian, Poisson, Binomial, …) \[y_i|\boldsymbol{x},\boldsymbol{\theta} \sim \pi(y_i|x_i,\boldsymbol{\theta}),\ i=1, \ldots, n\]
Latent Gaussian field \(\boldsymbol{x}|\boldsymbol{\theta}\sim N(\boldsymbol{\mu(\theta)},\boldsymbol{Q(\theta)}^{-1})\)
Hyperparameters (not necessarily Gaussian) \(\boldsymbol{\theta} \sim \pi(\boldsymbol{\theta})\)
Linear predictor accounts for effects of covariates in an additive way
\[\eta_i=\alpha+\sum_{k=1}^{n_{\beta}}\beta_k z_{ki}+\sum_{j=1}^{n_f}f^{(j)}(u_{ji})\]
\(\alpha\) intercept, \(\{\beta_k\}\)’s quantify the linear effects of covariates \(\{z_{ki}\}\) on response, \(\{f^{(j)}(\cdot)\}\)’s set of random effects defined in terms of some covariates \(\{u_{ji}\}\) (e.g. iid, rw1, ar1, CAR, …)
Latent Gaussian variables \(\boldsymbol{x}=(\alpha,\{\beta_k\},\{f^{(j)}\})|\boldsymbol{\theta}\sim N(\boldsymbol{\mu(\theta)},\boldsymbol{Q(\theta)}^{-1})\)
INLA
Compute posterior marginals for latent Gaussian field and hyperparameters
\[\pi(x_i|\boldsymbol{y})=\int \pi(x_i|\boldsymbol{\theta},\boldsymbol{y})\pi(\boldsymbol{\theta}|\boldsymbol{y})d\boldsymbol{\theta},\ \pi(\theta_j|\boldsymbol{y})=\int \pi(\boldsymbol{\theta}|\boldsymbol{y})d\boldsymbol{\theta}_{-j}\]
Use this form to construct nested approximations
\[\tilde \pi(x_i|\boldsymbol{y})=\int \tilde \pi(x_i|\boldsymbol{\theta},\boldsymbol{y})\tilde \pi(\boldsymbol{\theta}|\boldsymbol{y})d\boldsymbol{\theta},\ \tilde \pi(\theta_j|\boldsymbol{y})=\int \tilde \pi(\boldsymbol{\theta}|\boldsymbol{y})d\boldsymbol{\theta}_{-j}\]
This approximation can be integrated numerically with respect to \(\boldsymbol{\theta}\) \[\tilde \pi(x_i|\boldsymbol{y})=\sum_k \tilde \pi(x_i|\boldsymbol{\theta_k},\boldsymbol{y})\tilde \pi(\boldsymbol{\theta_k}|\boldsymbol{y})\times \Delta_k,\ \tilde \pi(\theta_j|\boldsymbol{y})=\sum_l \tilde \pi(\boldsymbol{\theta_l^*}|\boldsymbol{y})\times \Delta_l^*\]
\(\Delta_k\) area weight corresponding to \(\boldsymbol{\theta_k}\)
\(\Delta_l^*\) area weight corresponding to \(\boldsymbol{\theta_l^*}\)
INLA
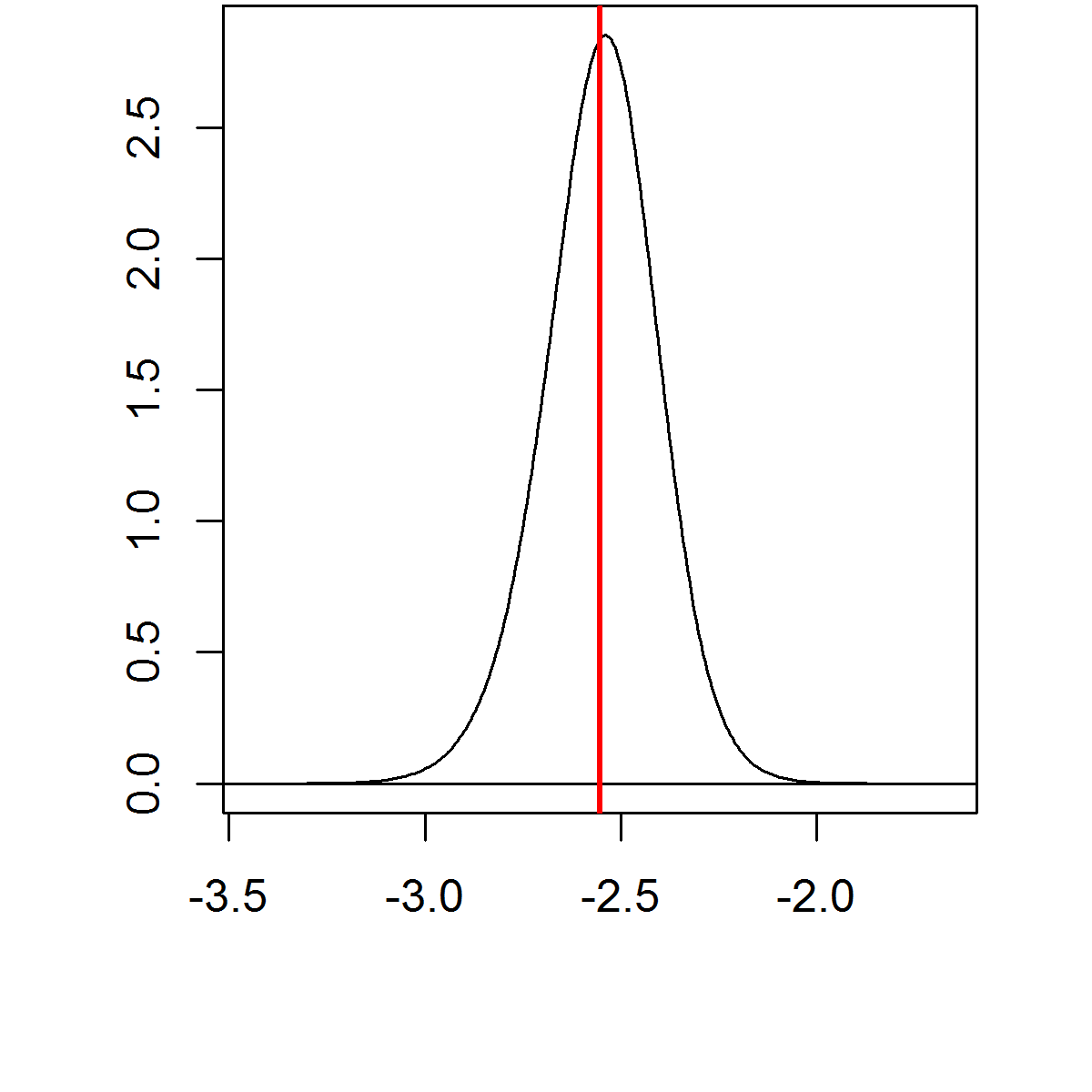
The approximated posterior distributions \(\tilde \pi (x_i|\boldsymbol{y})\) can be post-processed to compute quantities of interest like posterior expectations and quantiles
| Expectation | 95% C.I. |
|
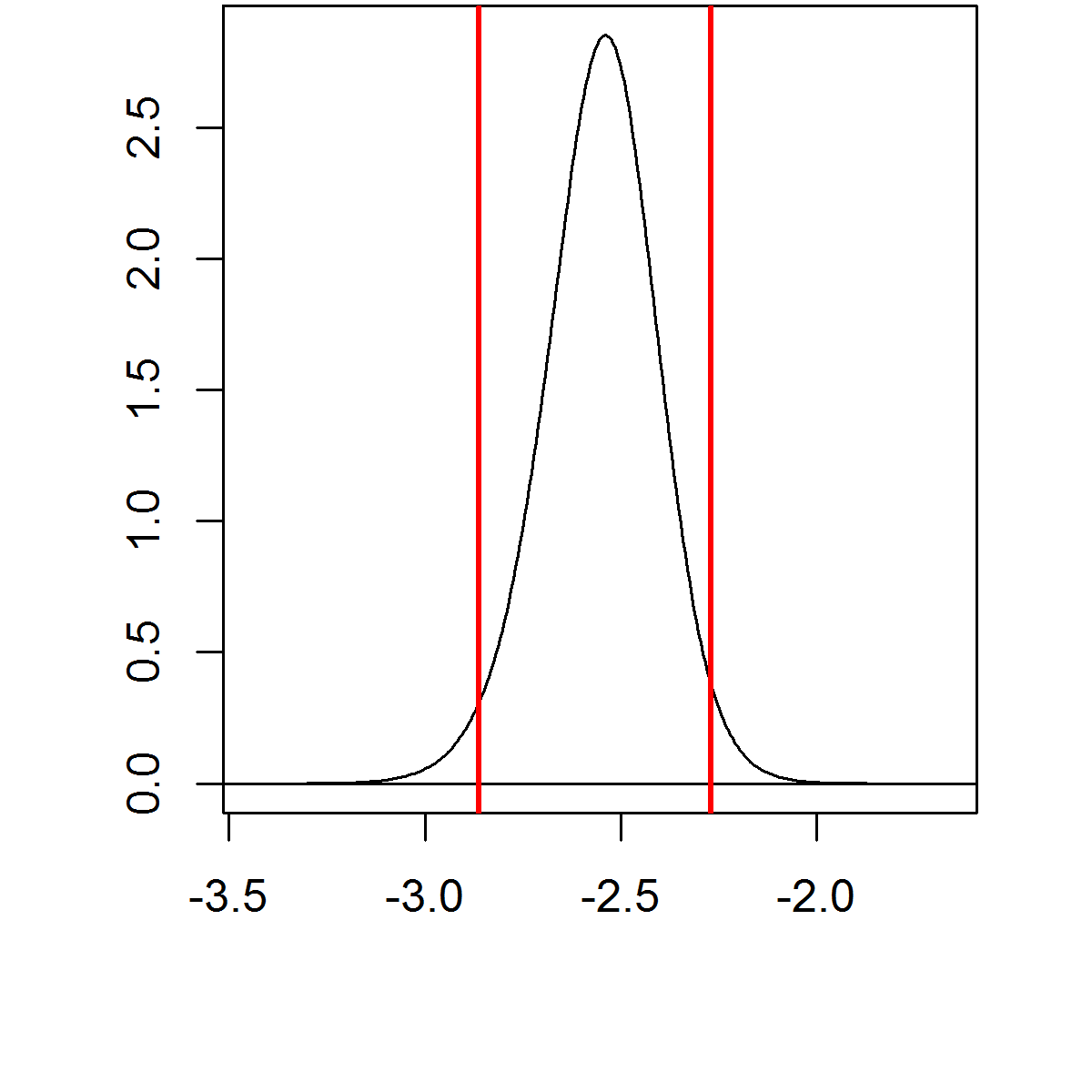
|
R-INLA package
Install INLA
R-INLA package
Model
\[Y_i | \eta_i, \sigma^2 \sim N(\eta_i, \sigma^2), i = 1, \ldots, n\] \[\eta_i = \beta_0 + \beta_1 \times x_{1i} + \beta_2 \times x_{2i} + u_i,\ u_i \sim N(0, \sigma^2_u)\]
- Write the linear predictor as a formula object in R
Random effects specified with f(). First argument id is an index vector that specifies the element of the random effect that applies to each observation, second argument model name
- Call the function
inla()
- Inspect the
resobject wich contains the fitted model.
Posteriors can be post-processed using a set of functions provided by R-INLA
Areal data
Areal data

Standardized Mortality Ratio (SMR) is often used to estimate disease risk
\[ SMR_i = \frac{Y_i}{E_i} = \frac{\mbox{number observed cases in area } i}{\mbox{number expected cases in area } i}\]
\(SMR_i = 1\) same number observed as expected
\(SMR_i > 1\) more observed than exp. (high risk)
\(SMR_i < 1\) less observed than exp. (low risk)
Example
\(SMR_i = \frac{Y_i}{E_i} = \frac{200}{100} = 2 > 1\) \(\rightarrow\) area \(i\) high risk (observed \(>\) expected) \(SMR_i = \frac{Y_i}{E_i} = \frac{100}{200} = 0.5 < 1\) \(\rightarrow\) area \(i\) low risk (observed \(<\) expected)
Indirect standardiz. to calculate expected cases
The expected number of cases in area \(i\) are the number of cases one would expect if population in area \(i\) behaved the way the standard pop. behaves
Standard population is considered as the whole population (all areas)
Population is stratified by several factors (e.g., age and gender)
\(r_j^{(std)}=\frac{\mbox{number cases}}{\mbox{population}}\) rate in stratum \(j\) in the standard population
\(n_j^{(i)}\): population in stratum \(j\) of area \(i\)
Limitations
SMRs easy to calculate but may be misleading and unreliable in areas with small populations or rare diseases. Models enable to incorporate covariates & borrow information from neighboring areas to obtain smoothed relative risks
Areal models
Model to estimate disease relative risk \(\theta_i\) in areas \(i=1,\ldots,n\)
\[Y_i|\theta_i \sim Poisson(E_i \times \theta_i)\] \[\log(\theta_i) = \boldsymbol{z}_i \boldsymbol{\beta} + u_i + v_i\]
- \(Y_i\) observed cases, \(E_i\) expected cases, \(\theta_i\) relative risk in area \(i\)
Fixed effects quantify the effects of the covariates on the disease risk
- \(\boldsymbol{z}_i = (1, z_{i1}, \ldots, z_{ip})\) intercept and cov. \(\beta = (\beta_0, \beta_1, \ldots, \beta_p)'\) coeff.
Random effects represent residual variation not explained by the covariates
- \(u_i\): structured spatial effect to account for spatial dependence between relative risks (close areas show more similar risk than not close areas)
- \(v_i\): unstructured effect to account for independent noise
Tutorial
Modeling areal data (lung cancer risk in Pennsylvania, USA)
https://www.paulamoraga.com/book-spatial/disease-risk-modeling.html
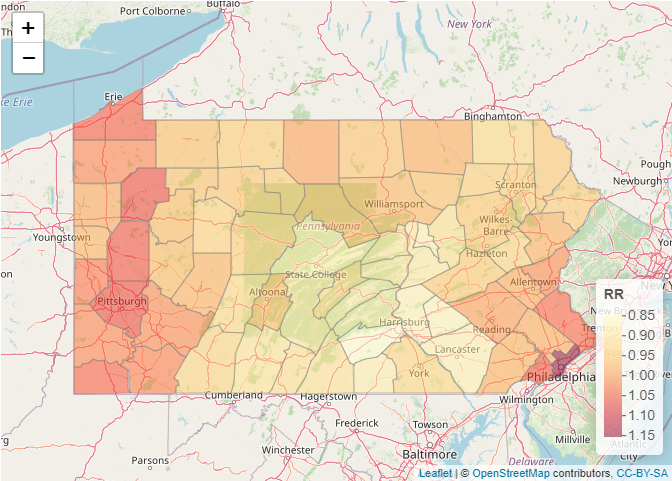
Geostatistical data
Geostatistical data

Geostatistical models
Models to predict prevalence at unsampled locations
\[Y_i|P(\boldsymbol{x}_i)\sim \mbox{Binomial} (n_i, P(\boldsymbol{x}_i))\] \[\mbox{logit}(P(\boldsymbol{x}_i)) = \boldsymbol{z}_i \boldsymbol{\beta} + S(\boldsymbol{x}_i) + u_i\]
\(Y_i\) number people positive, \(n_i\) number people tested, \(P(\boldsymbol{x}_i)\) prevalence at \(\boldsymbol{x}_i\)
Covariates based on characteristics known to affect disease transmission (temperature, precipitation, vegetation, elevation, land cover, population density, etc.)
Random effects model residual variation not explained by covariates

Inference using INLA and SPDE
Integrated nested Laplace approximations (INLA) is a computational approach to perform approximate Bayesian inference in latent Gaussian models
In the Stochastic partial differential equation (SPDE) approach, the continuously indexed Gaussian random field \(S\) is represented as a discretely indexed Gaussian Markov random field (GMRF) by means of a finite basis function defined on a triangulation of the study region
\[S(\boldsymbol{x}) = \sum_{g=1}^G \psi_g(\boldsymbol{x}) S_g\]
\(\psi_g(\cdot)\) piecewise polynomial basis functions on each triangle
\(\{S_g \}\) zero-mean Gaussian distributed
\(G\) number of vertices in triangulation
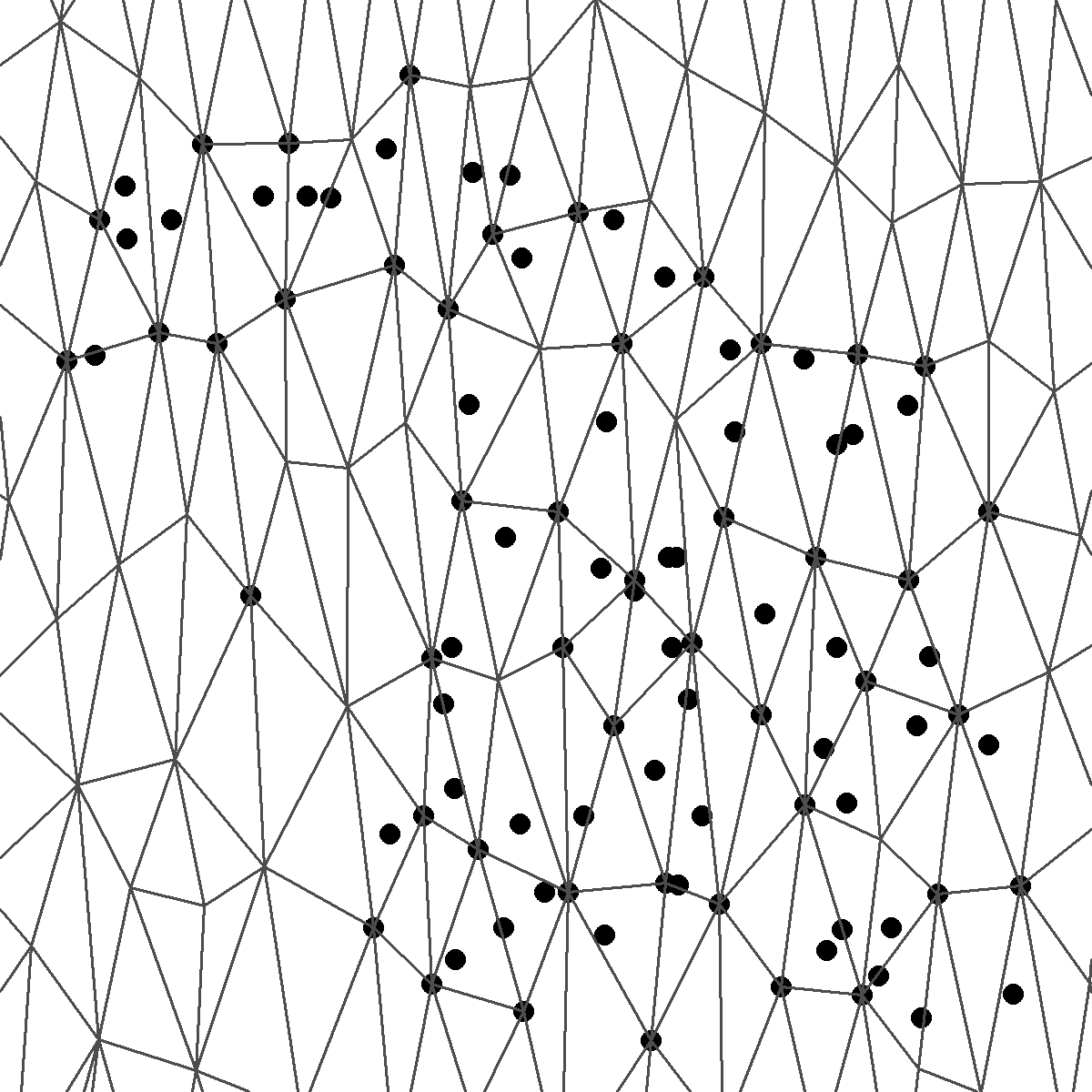
Projection matrix
\(S(\boldsymbol{x})\) weighted average of the GMRF values at the vertices of the triangle containing the point. Weights = barycentric coordinates
\[S(\boldsymbol{x}) \approx \frac{T_{1}}{T}S_1 + \frac{T_{2}}{T}S_2 + \frac{T_{3}}{T}S_3\]
\(T_1, T_2, T_3\) areas subtriangles formed by \(\boldsymbol{x}\) and vertices. \(T\) area whole triangle
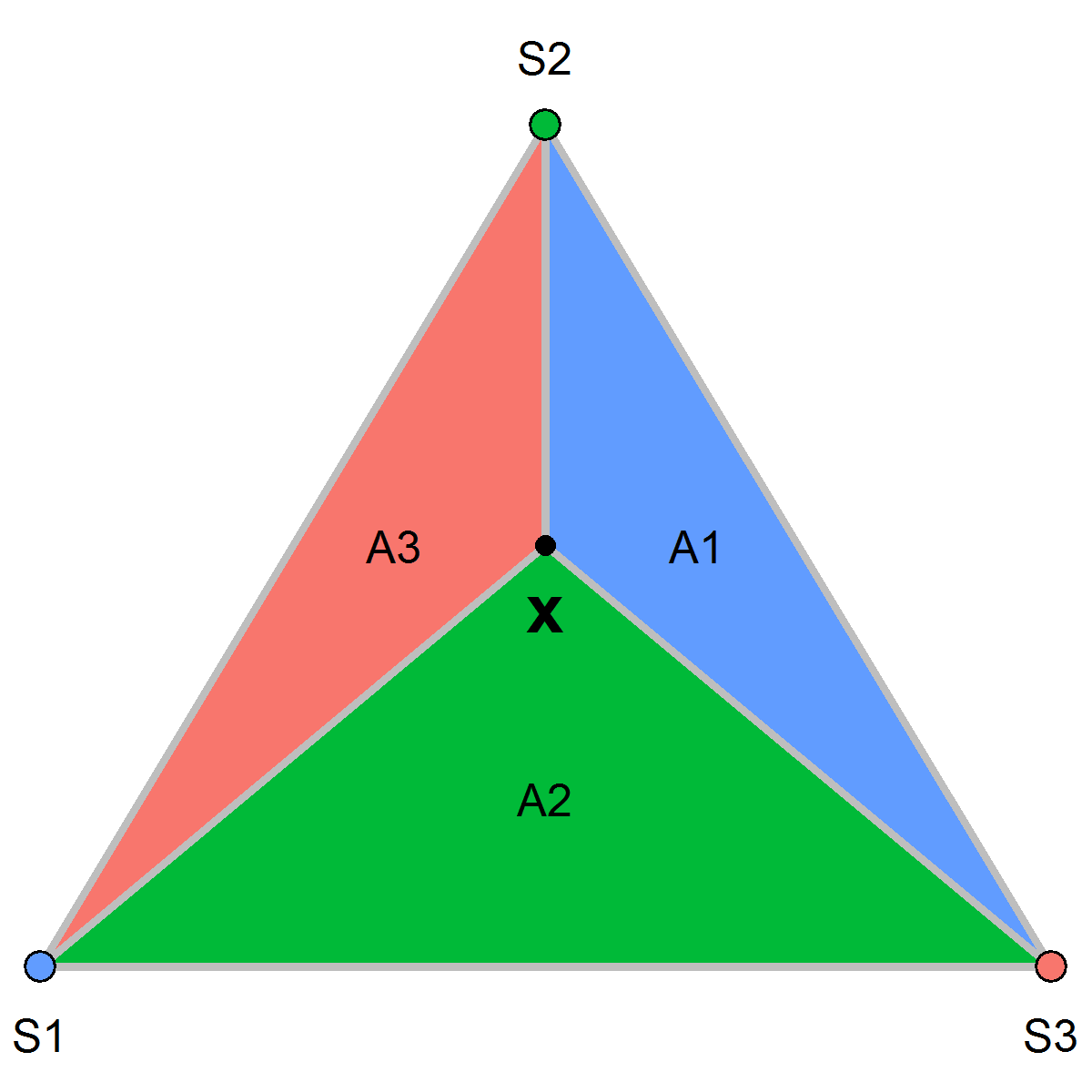
Projection matrix
\[S(\boldsymbol{x}_i) \approx \sum_{g=1}^G A_{ig} S_g\] \(A\) projection matrix that maps GMRF from observations to triangulation nodes
Row \(i\) corresponding to observation \(i\) has possibly three non-zero values at columns that represent the vertices of the triangle containing the point
(non-zero values are equal to the barycentric coordinates)
\[A = \begin{bmatrix} A_{11} & A_{12} & A_{13} & \dots & A_{1G} \\ A_{21} & A_{22} & A_{23} & \dots & A_{2G} \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ A_{n1} & A_{n2} & A_{n3} & \dots & A_{nG} \end{bmatrix} = \begin{bmatrix} 1 & 0 & 0 & \dots & 0 \\ 0 & 0 & 1 & \dots & 0 \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 0.2 & 0.2 & 0 & \dots & 0.6 \end{bmatrix}\]
Tutorial
Modeling geostatistical data (air pollution in USA)
https://www.paulamoraga.com/book-spatial/sec-geostatisticaldataSPDE.html
Tutorial
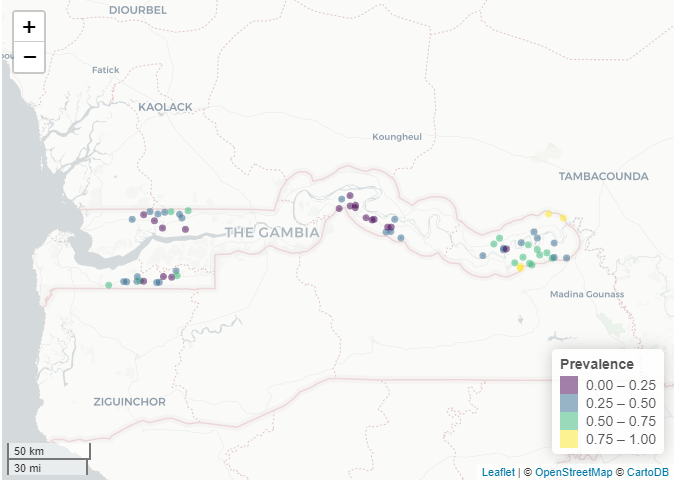
Modeling geostatistical data (malaria prevalence in The Gambia)
https://www.paulamoraga.com/book-geospatial/sec-geostatisticaldataexamplespatial.html
Point patterns
Point patterns

Assume point pattern \(\{s_i: i=1, \ldots, n\}\) has been generated as a realization of a point process \(Z = \{Z(s): s \in \mathbb{R}^2\}\)
A point process model can be used to estimate the intensity of events, identify patterns in the distribution of the observed locations, and learn about the correlation between the locations and spatial covariates
Intensity of the process
Intensity of the process: mean number of events per unit area at location \(s\) \[\lambda(s) = lim_{|ds|\rightarrow 0} \frac{E[N(ds)]}{|ds|}\]

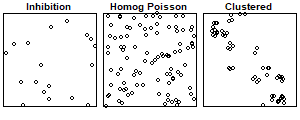
Complete Spatial Randomness
Point processes are stochastic models that describe the locations of events of interest and possibly some additional information such as marks that inform about different types of events (e.g., cases and controls)
Point processes provide models for point patterns, with complete spatial randomness (CSR) being the simplest theoretical model
CSR helps differentiate between regular and clustered patterns

Homogeneous Poisson process (CRS)
Simple model that assumes events are equally likely to occur at any location within the study area, independent of the locations of other events
- the number of events in any region \(A\) follows a Poisson distribution with mean \(\lambda |A|\), where \(\lambda\) is a constant value denoting the intensity and \(|A|\) is the area of \(A\)
- given that there are \(n\) events inside \(A\), the locations of these events are independent and uniformly distributed in \(A\)

Log-Gaussian Cox process (LGCP)
Log-Gaussian Cox processes (LGCP) are typically used to model phenomena that are environmentally driven
A LGCP is a Poisson process with a varying intensity which is itself a stochastic process of the form \[\Lambda(s) = exp(\eta(s)),\] where \(\eta = \{\eta(s): s \in \mathbb{R}^2\}\) is spatial Gaussian process
Conditional on \(\Lambda(s)\), the point process is a Poisson process with intensity \(\Lambda(s)\) 1. the number of events in any region \(A\) is distributed as \(Poisson(\int_A \Lambda(s)ds)\) 2. the locations of events are an independent random sample with probability density proportional to \(\Lambda(s)\)
Fitting a log-Gaussian Cox process model
Assume point pattern \(\{s_i: i=1, \ldots, n\}\) has been generated as a realization of a LGCP with intensity \(\Lambda(s)= exp(\eta(s))\)
A LGCP can be fitted by discretizing the study region into a grid with \(n_1 \times n_2\) cells \(\{s_{ij}: i=1,\ldots,n_1, j=1,\ldots,n_2\}\). \(|s_{ij}|\) area of cell \(s_{ij}\)
Mean number of events in cell \(s_{ij}\):
\(\Lambda_{ij}=\int_{s_{ij}} exp(\eta(s))ds \approx |s_{ij}| exp(\eta_{ij})\)
\(y_{ij}|\eta_{ij} \sim Poisson(|s_{ij}| exp(\eta_{ij}))\)
\(\eta_{ij} = \beta_0 + \beta_1 \times cov(s_{ij}) + f_s(s_{ij}) + f_u(s_{ij})\)

- \(y_{ij}\) observed number of locations in grid cell \(s_{ij}\)
- \(\beta_0\) intercept, \(cov(s_{ij})\) covariate at \(s_{ij}\), \(\beta_1\) coefficient of \(cov(s_{ij})\)
- \(f_s()\) spatially structured random effect (CAR model on a regular lattice)
- \(f_u()\) unstructured random effect
Tutorial
Modeling point patterns (sloths occurrence in Costa Rica)
http://www.paulamoraga.com/tutorial-point-patterns
spocc package: https://docs.ropensci.org/spocc/

Reproducible documents with R Markdown
R Markdown
R Markdown can be used to turn our analysis into fully reproducible documents that can be shared with others. Output formats include HTML, PDF or Word. An R Markdown file is written with Markdown syntax with embedded R code, and can include narrative text, tables and visualizations
http://www.paulamoraga.com/book-geospatial/sec-rmarkdown.html

YAML header
Markdown syntax
**bold text**bold text
*italic text*italic text
- unordered item
- unordered item- unordered item
- unordered item
1. first item
2. second item- first item
- second item
# First-level header
## Second-level header
### Third-level headerFirst-level header
Second-level header
Third-level header
$$\int_0^\infty e^{-x^2} dx=\frac{\sqrt{\pi}}{2}$$\[\int_0^\infty e^{-x^2} dx=\frac{\sqrt{\pi}}{2}\]
R code chunks
Options:
echo=FALSEcode will not be shown in the document, but it will run and the output will be displayed in the documenteval=FALSEcode will not run, but it will be shown in the documentinclude=FALSEcode will run, but neither the code nor the output will be included in the documentresults='hide'output will not be shown, but the code will run and will be displayed in the documentcache=TRUEcode chunk is not executed if it has been executed before and nothing in the code chunk has changed since thenerror=FALSE,warning=FALSE,message=FALSEsupress errors, warnings or messages
R Markdown

Interactive dashboards with flexdashboard
Dashboards with flexdashboard
The R package flexdashboard uses R Markdown to publish a group of related data visualizations as a dashboard
http://www.paulamoraga.com/book-geospatial/sec-flexdashboard.html

Layout

Dashboards with flexdashboard

Tutorial
Interactive dashboards with flexdashboard to communicate results
https://www.paulamoraga.com/book-geospatial/sec-flexdashboard
Example (lung cancer risk in Pennsylvania, USA)
http://www.paulamoraga.com/tutorial-flexdashboard-example

Shiny web applications
Shiny web applications
Shiny is a web application framework for R that enables to build interactive web applications
Examples: Gallery, Example 1, Example 2
Resources: RStudio tutorial, Mastering Shiny Book, Cheatsheet
SpatialEpiApp
SpatialEpiApp is a Shiny app for disease risk estimation, cluster detection, and interactive visualization
- Risk estimates by fitting Bayesian models with INLA
- Detection of clusters by using the scan statistics in SaTScan

Structure of a Shiny App
A Shiny app can be built by creating a directory (called, for example, appdir) that contains an R file (called, for example, app.R) with three components:
uiuser interface object which controls layout and appearance of the appserver()function with instructions to build objects displayed in theui- call to
shinyApp()that creates the Shiny app from theui/serverpair
Save app.R inside directory appdir. Launch the app:
Directory appdir can also contain data or other R scripts needed by the app. We can also write two separate files ui.R and server.R. This permits an easier management of the code for large apps
Inputs
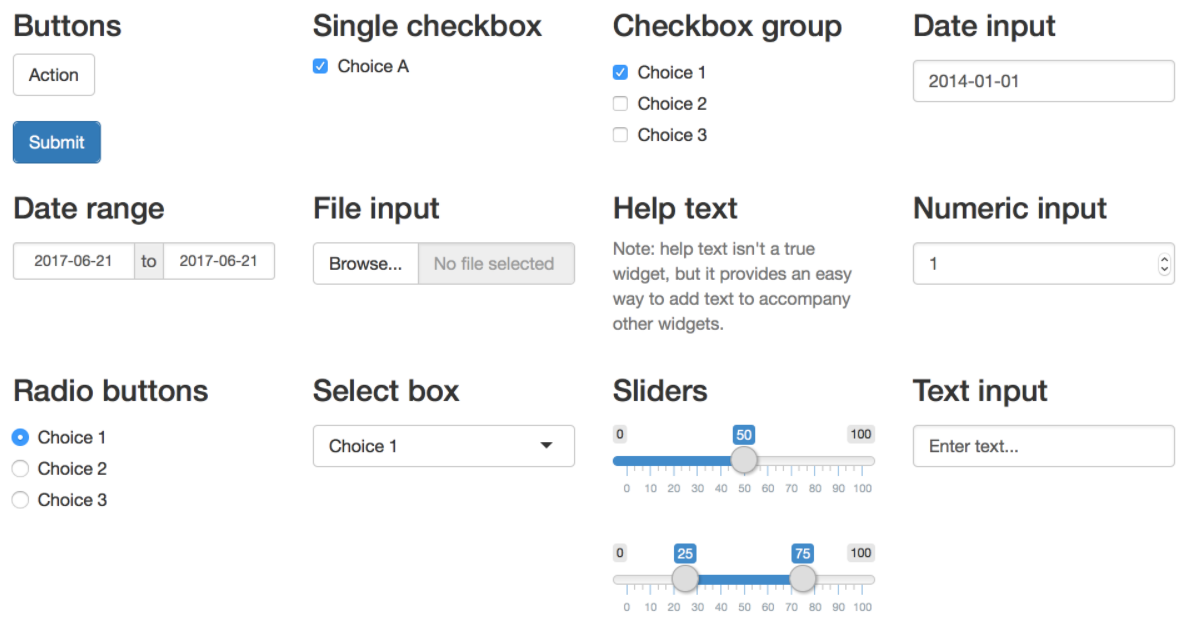
Outputs
Plots, tables, texts, images
Inputs, outputs and reactivity
Inputs: we can interact with the app by modifying their values
Outputs: objects we want to show in the app
ui <- fluidPage(
*Input(inputId = myinput, label = mylabel, ...)
*Output(outputId = myoutput, ...)
)
server <- function(input, output){
output$myoutput <- render*({
# code to build the output.
# If it uses an input value (input$myinput),
# the output will be rebuilt whenever the input value changes
})}We can include inputs by writing an *Input() function in the ui
(e.g., textInput(), dateRangeInput(), fileInput())
Outputs can be created by writing an *Output() function in the ui, and the R code to build the output inside a render*() function in server() (e.g., textOutput() and renderText(), tableOutput() and renderTable())
fluidPage() creates a display that automatically adjusts the app to the dimensions of the browser window
Inputs, outputs and reactivity
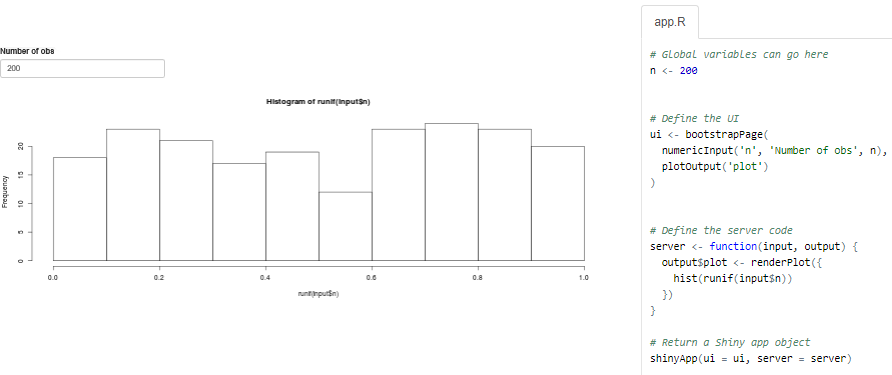
https://shiny.rstudio.com/gallery/single-file-shiny-app.html
Sharing a Shiny app
Option 1: Share R scripts with other users
- users need R
Option 2: Host app as a web page at its own URL
- users do not need R
- app can be navigated through the internet with a web browser
- host apps on own servers or using one of the ways RStudio offers such as shinyapps.io and Shiny Server
Shiny practical 1
Inputs, outputs and reactivity
Inputs: we can interact with the app by modifying their values
Outputs: objects we want to show in the app
ui <- fluidPage(
*Input(inputId = myinput, label = mylabel, ...)
*Output(outputId = myoutput, ...)
)
server <- function(input, output){
output$myoutput <- render*({
# code to build the output.
# If it uses an input value (input$myinput),
# the output will be rebuilt whenever the input value changes
})}We can include inputs by writing an *Input() function in the ui
(e.g., textInput(), dateRangeInput(), fileInput())
Outputs can be created by writing an *Output() function in the ui, and the R code to build the output inside a render*() function in server() (e.g., textOutput() and renderText(), tableOutput() and renderTable())
fluidPage() creates a display that automatically adjusts the app to the dimensions of the browser window
Example
Shiny app that greets the user by name
Shiny widgets
Gallery: https://shiny.posit.co/r/gallery/widgets/widget-gallery/
List of inputs and outputs: https://shiny.posit.co/r/reference/shiny/latest/
Exercise
Build a Shiny app containing the following inputs and outputs:
Slider input with label
"Select dates:", minimum date"2020-01-01", maximum date"2020-12-31", and initial value"2020-01-31".Slider input to select a value between 0 and 100 where the interval between each selectable value on the slider is 5. Add animation so when the user presses play the input widget scrolls through automatically.
Output with a text showing the input values selected.
Exercise
library(shiny)
ui <- fluidPage(
sliderInput("date", "Select date:",
min = as.Date("2020-01-01"),
max = as.Date("2020-12-31"),
value = as.Date("2020-01-31")),
sliderInput("number", "Select a number:",
min = 0, max = 100, value = 0,
step = 5, animate = TRUE),
textOutput("inputvaluesselected")
)
server <- function(input, output, session) {
output$inputvaluesselected <- renderText({
paste("Date", input$date, ",",
"number", input$number, ".")
})
}
shinyApp(ui, server)Reactive expressions
Reactive expressions take reactive values or other reactive expressions, and return an object to be used in the app
Reactive expressions
Fibonacci sequence: 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, …
# Function to calculate nth number in Fibonacci sequence
fib <- function(n){ifelse(n<3, 1, fib(n-1)+fib(n-2))}
ui <- fluidPage(
numericInput("n", "Enter number", value = 1),
textOutput("nthvalue"),
textOutput("nthvalueInv")
)
server <- function(input, output) {
# time consuming calculation that we only want to do once
# do calculation in a reactive expression
currentFib <- reactive({fib(as.numeric(input$n))})
output$nthvalue <- renderText({currentFib()})
output$nthvalueInv <- renderText({1/currentFib()})
}
shinyApp(ui, server)Observers
Observers, like reactive expressions, can take reactive values and reactive expressions. However, they do not return any values. Instead of returning values they have side effects
ui <- fluidPage(
sliderInput("controller", "Controller", 0, 20, 10),
textInput("inText", "Input text"),
textInput("inText2", "Input text 2")
)
server <- function(input, output, session) {
observe({
# We use input$controller multiple times so we save it as x
x <- input$controller
# changes the value of input$inText
updateTextInput(session, "inText", value = paste("New text", x))
# changes the value and label of input$inText2
updateTextInput(session, "inText2",
label = paste("New label", x),
value = paste("New text", x))
})
}
shinyApp(ui, server)Events
Sometimes we may want to wait for a specific action to be taken from the user, like clicking an actionButton() or an actionLink(), before calculating an expression or taking an action
An event is a reactive value or expression that is used to trigger other calculations in this way
observeEvent() and eventReactive() can be used for event handling:
observeEvent()to perform an action in response to an eventeventReactive()to create a calculated value that only updates in response to an event
Events
ui <- fluidPage(
numericInput("number", "Value", 5),
actionButton("button", "Show"),
tableOutput("table")
)
server <- function(input, output) {
# observeEvent() and eventReactive()
# take a dependency on input$button, but
# not on any of the stuff inside the functions
# Take an action every time button is pressed
# Here, print a message to the console
observeEvent(input$button, {cat("Showing",input$number,"rows\n")})
# Calculate df every time button is pressed
df <- eventReactive(input$button, {head(cars, input$number)})
output$table <- renderTable({df()})
}
shinyApp(ui, server)Uploads
ui <- fluidPage(
fileInput("upload", label = "Select file",
buttonLabel = "Upload...", multiple = TRUE),
tableOutput("files")
)
server <- function(input, output, session) {
output$files <- renderTable(input$upload)
}
shinyApp(ui, server)fileInput() returns a data frame with four columns: name, size, type, datapath
Uploads
input$upload initialized to NULL on page load. Need req(input$upload) to make sure code waits until first file is uploaded
accept argument limits the possible inputs. But it is only a suggestion to browser not always enforced. Good practice validate (tools::file_ext())
ui <- fluidPage(
fileInput("upload", "Select file", accept = c(".csv", ".tsv")),
numericInput("n", "Rows", value = 5, min = 1, step = 1),
tableOutput("head")
)
server <- function(input, output, session) {
data <- reactive({
req(input$upload)
ext <- tools::file_ext(input$upload$name)
switch(ext, # file extension excluding leading dot
csv = vroom::vroom(input$upload$datapath, delim = ","),
tsv = vroom::vroom(input$upload$datapath, delim = "\t"),
validate("Invalid file; Please upload a .csv or .tsv file"))
})
output$head <- renderTable({head(data(), input$n)})
}
shinyApp(ui, server)Downloads
In ui, downloadButton(id) or downloadLink(id) so user can click to download a file. In server(), downloadHandler() with two fn arguments
ui <- fluidPage(
selectInput("dataset", "Pick a dataset", ls("package:datasets")),
tableOutput("preview"),
downloadButton("download", "Download .tsv")
)
server <- function(input, output, session) {
data <- reactive({
out <- get(input$dataset, "package:datasets")
if(!is.data.frame(out)){
validate(paste0("'", input$dataset, "' is not a data frame"))}
out
})
output$preview <- renderTable({head(data())})
output$download <- downloadHandler(
# returns a file name shown in the download dialog box
filename = function(){paste0(input$dataset, ".tsv")},
# argument file is the path to save the file. Saves the file
# in a place Shiny knows about, so it can send it to the user
content = function(file){vroom::vroom_write(data(), file)})
}
shinyApp(ui, server)Appearance
Customize apps with the bslib package https://rstudio.github.io/bslib/
Bootswatch themes: https://bootswatch.com/
Preview themes with bslib::bs_theme_preview()
Customize app
In ui, pick a bootswatch theme to change the overall look of the Shiny app
Customize plots
In the server() function, call
to customize ggplot2, lattice and base plots to match the style of the app. This automatically determines all of the settings from the app theme
Appearance
library(ggplot2)
ui <- fluidPage(
theme = bslib::bs_theme(bootswatch = "darkly"),
sidebarLayout(
sidebarPanel(
textInput("txt", "Text input:", "text here"),
sliderInput("slider", "Slider input:", 1, 100, 30)),
mainPanel(
plotOutput("plot"))
)
)
server <- function(input, output, session){
thematic::thematic_shiny()
output$plot <- renderPlot({
ggplot(mtcars, aes(wt, mpg)) + geom_point() + geom_smooth()
})
}
shinyApp(ui, server)Layout
https://shiny.posit.co/r/articles/build/layout-guide/
Laying out the components of an application:
fluidPage()creates a display that automatically adjusts to the dimensions of the browser window- Sidebar layout:
titlePanel()andsidebarLayout(sidebarPanel(), mainPanel()).sidebarPanel()for inputs &mainPanel()for outputs - Grid layout:
fluidRow(column(), column(), ...). Column widths should add up to 12 within afluidRow()container - Tabsets:
tabsetPanel()for presenting the components using tabs - Navlists:
navlistPanel()for presenting the components as a sidebar list - Navbar pages:
navbarPage()multi-page user interface with navigation bar
Exercise: Build a Shiny app with a grid layout that contains a row with two columns containing two plots, each column taking half the app
Exercise
Shiny practical 2
Leaflet
https://rstudio.github.io/leaflet/shiny.html
library(leaflet)
ui <- fluidPage(
leafletOutput("map"),
actionButton("button", "New locations")
)
server <- function(input, output, session){
# when button is pressed, generate 40 new locations
points <- eventReactive(input$button, {cbind(rnorm(40)*2+13, rnorm(40)+48)}, ignoreNULL = FALSE)
output$map <- renderLeaflet({
leaflet() %>% addTiles() %>%
addMarkers(data = points())
})
}
shinyApp(ui, server)Set ignoreNULL = FALSE to initially perform the calculation and let the user recalculate (reference)
Leaflet
This works, but reactive inputs and expressions that affect the renderLeaflet() expression will cause the entire map to be redrawn from scratch and reset the map position and zoom level
Use leafletProxy() instead of leaflet() to have more control over the map, such as changing the color of a single polygon or adding a marker at the point of a click without redrawing the entire map
Typically, use leaflet() to create the static aspects of the map, and leafletProxy() to modify the dynamic elements of a map that is already running in the page
Leaflet
library(leaflet)
ui <- fluidPage(
leafletOutput("map"),
actionButton("button", "New locations")
)
server <- function(input, output, session){
# when button is pressed, generate 40 new locations
points <- eventReactive(input$button,
{cbind(rnorm(40)*2+13, rnorm(40)+48)}, ignoreNULL = FALSE)
# leaflet() to include aspects that do not change in the map
output$map <- renderLeaflet({
leaflet() %>% addTiles() %>%
setView(lng = 15, lat = 48, zoom = 6)
})
# leafletproxy() to include aspects that change in the map
observe({
leafletProxy("map") %>%
clearMarkers() %>% addMarkers(data = points())
})
}
shinyApp(ui, server)Shiny practical 3
Interactive dashboards with
flexdashboard and Shiny

Interactive dashboards with
flexdashboard and Shiny
Flexdashboard
https://www.paulamoraga.com/book-geospatial/sec-flexdashboard
Flexdashboard and Shiny
https://www.paulamoraga.com/book-geospatial/sec-dashboardswithshiny
Add runtime: shiny to the YAML header
Add a column on the left-hand side of the dashboard to add the input controls. Add .{sidebar} to have a special background color
Add Shiny inputs and outputs as appropriate
Include visualizations by wrapping them in a call to render*({}) to dynamically respond to changes
Leaflet map with leafletProxy()
https://www.paulamoraga.com/book-geospatial/sec-dashboardswithshiny
# leaflet() to include aspects that do not change in the map
output$map <- renderLeaflet({
leaflet() %>% addTiles() %>% setView(lng = 0, lat = 30, zoom = 2)
})
# leafletproxy() to include aspects that change in the map
observe({
if (nrow(mapFiltered()) == 0) {return(NULL)}
leafletProxy("map", session, data = mapFiltered()) %>%
clearShapes() %>% clearControls() %>%
addPolygons(fillColor = ~ pal(PM2.5), color = "white",
fillOpacity = 0.7, label = ~labels,
highlight = highlightOptions(color = "black",
bringToFront = TRUE)) %>%
leaflet::addLegend(pal = pal, values = ~PM2.5, opacity = 0.7, title = "PM2.5")
})
leafletOutput("map")Interactive dashboards with shinyflexdashboard
Package shinydashboard provides another way to create dashboards with Shiny
Building a Shiny app to upload and visualize spatio-temporal data
https://www.paulamoraga.com/book-geospatial/sec-shinyexample.html
Shiny practical 4
Sharing Shiny apps with shinyapps.io
Deploy a Shiny app:
https://docs.rstudio.com/shinyapps.io/getting-started.html
Create a shinyapps.io account: https://www.shinyapps.io/
Configure rsconnect
library(rsconnect)
rsconnect::setAccountInfo(name = "<ACCOUNT>", token = "<TOKEN>",
secret = "<SECRET>")Deploy Shiny app
Before deployApp(), use setwd() to set the working directory to the folder where app.R is. Or use the argument appDir of deployApp() to set the folder where app.R is
Join my research group at KAUST!
KAUST is an international university located on the shores of the Red Sea
All students receive a living allowance, free housing and medical coverage
👩💻 Potential research areas include the development of innovative statistical and computational methods for health and environmental applications
💪 Work closely with collaborators at KAUST and around the world
✈️ Generous travel funding for conferences and collaborative work
✨ Excellent research environment. Superb equipment and research facilities
References
Ribeiro Amaral, A., et al. (2022). Spatio-temporal modeling of infectious diseases by integrating compartment and point process models. SERRA, 37, 1519-1533
Moraga, P. and Baker, L. (2022). rspatialdata: a collection of data sources and tutorials on downloading and visualising spatial data using R. F1000Research, 11:77
Moraga, P., et al. (2019). epiflows: an R package for risk assessment of travel-related spread of disease. F1000Research, 7:1374
Moraga, P. (2018). Small Area Disease Risk Estimation and Visualization Using R. The R Journal, 10(1):495-506
Moraga, P. (2017). SpatialEpiApp: A Shiny Web Application for the analysis of Spatial and Spatio-Temporal Disease Data. Spatial and Spatio-temporal Epidemiology, 23:47-57
Moraga, P., et al. (2017). A geostatistical model for combined analysis of point-level and area-level data using INLA and SPDE. Spatial Statistics, 21:27-41
Moraga, P. and Kulldorff, M. (2016). Detection of spatial variations in temporal trends with a quadratic function. Statistical Methods for Medical Research, 25(4):1422-1437
Hagan, J. E., Moraga, P., et al. (2016). Spatio-temporal determinants of urban leptospirosis transmission: Four-year prospective cohort study of slum residents in Brazil. PLOS Neglected Tropical Diseases, 10(1): e0004275
Moraga, P., et al. (2015). Modelling the distribution and transmission intensity of lymphatic filariasis in sub-Saharan Africa prior to scaling up interventions: integrated use of geostatistical and mathematical modelling. Parasites & Vectors, 8:560



