R packages
In this tutorial we provide an introduction on how to create R packages. Resources that have been used to write this tutorial and that can be read to better understand and expand on the material covered here are the following:
R packages https://r-pkgs.org/
R package primer: https://kbroman.org/pkg_primer/
Mastering Software Development in R: https://bookdown.org/rdpeng/RProgDA/
Writing R extensions: https://cran.r-project.org/doc/manuals/R-exts.html
Package development: https://support.rstudio.com/hc/en-us/sections/200130627-Package-Development
Package websites with
pkgdown: https://pkgdown.r-lib.org/index.htmlrOpenSci: https://ropensci.org/
rOpenSci Packages: Development, Maintenance, and Peer Review: https://devguide.ropensci.org/
rOpenSci Statistical Software Peer Review: https://stats-devguide.ropensci.org/
1 R packages
R packages provide a way to distribute R code, data and documentation. R packages can be shared for reuse by others in several ways. For example, they can be contributed to the Comprehensive R Archive Network (CRAN), put in GitHub, or distributed privately using files shares.
R packages are directories with subdirectories containing R functions, data, documentation and other information. The minimal requirements for an R package are the following:
- Subdirectory
Rthat contains R files with functions - Subdirectory
manthat contains the documentation DESCRIPTIONfile with metadata for the package such as the name, version number and authorNAMESPACEfile that specifies exported functions that can be accessed by the users and imported functions from other packages- Other common parts include subdirectories
data,testsandvignettes
Package structure and state
https://r-pkgs.org/package-structure-state.html
The contents of a package can be stored as
- source: directory with subdirectories as above
- bundle: single compressed file (
.tar.gz) - binary: single compressed file optimized for a specific OS
Exercise Inspect the source of the
ggplot2 package in GitHub: https://github.com/tidyverse/ggplot2
Exercise Inspect the ggplot2 package on
CRAN: https://cran.r-project.org/web/packages/ggplot2/index.html
Exercise Find the folder where R packages are
installed in your computer and inspect the components of one of the
packages. You can use .libPaths() to obtain the path where
R packages are installed.
Building a package
To create an R package, we can use the devtools and usethis packages
which include a variety of tools aimed at package development. The roxygen2 package
allows us to easily create documentation for the functions and data
contained in the packages. The following functions are key for package
development.
usethis::create_package(): creates the file structure for a new packageusethis::use_r(): creates R filesdevtools::load_all(): loads all functions in a package like when a package is installed and attached withlibrary()usethis::use_package(): adds a package dependency to theDESCRIPTIONfileusethis::use_data(): saves an object in the R session as a dataset in the packagedevtools::document(): creates documentation files inman/and theNAMESPACEfile from roxygen2 code (it is a wrapper ofroxygen2::roxygenize())usethis::use_vignette(): creates a new vignette invignettes/devtools::check(): builds and checks a package for any ERRORs, WARNINGs, or NOTEsdevtools::install(): usesR CMD INSTALLto install a package

2 Creating an R package: a small example
Here we provide a small example on how to create an R package called
mypackage that has a function called
fnAreaCircle() that calculates the area of a circle given
its radius.
To develop the package, we use the devtools package
(which also uses other packages such as usethis for package
development).
To start the development of a new R package, we attach the
devtools package and call the create_package()
function that initializes the components of the package.
library(devtools)
usethis::create_package("C://mypackage")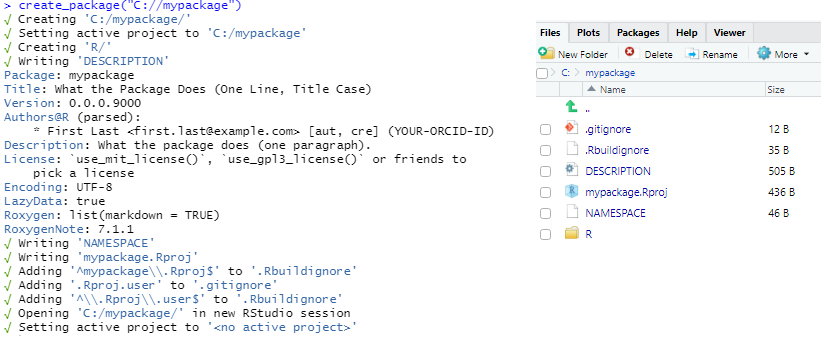
This creates the following files:
- Folder
R DESCRIPTIONNAMESPACEmypackage.Rproj.Rbuildignorelists files that should not be included when building the R package.gitignorelist files that Git should ignore such as some files created by R and RStudio
We can add content and modify the R package created. We start by
creating R functions in .R files and saving them in the
R/ subdirectory of the package. We can use the
use_r() function to create and/or open a file in
R/.
Here, we create fnAreaCircle.R by using
use_r("fnAreaCircle"). Then, we edit the file by adding the
function fnAreaCircle() that calculates the area of a
circle by passing its radius.
use_r("fnAreaCircle")fnAreaCircle <- function(r){
area <- pi*r^2
return(area)
}We can test the R functions created by calling
load_all(). load_all() makes the R functions
available for use, as when we attach the package via
library().
load_all()
fnAreaCircle(2)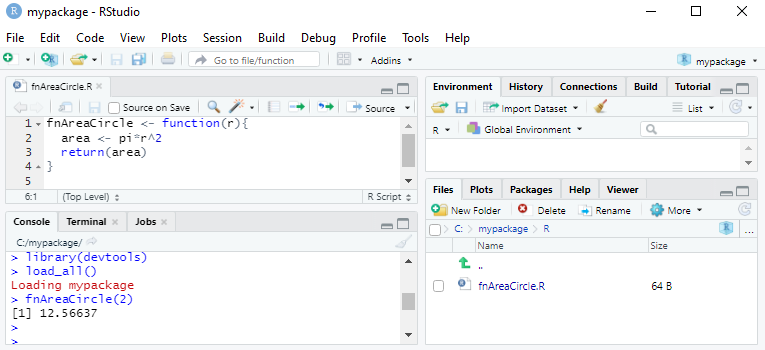
Then, we can use check() to check whether the full
package works.
check()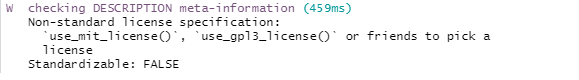
The output of check() shows a Warning because a license
for the package was not chosen. For this example, let us choose the MIT
license. We run use_mit_license() and this creates
LICENSE.md. Then, we run check() again.
use_mit_license()
check()
Since the output of check() shows no errors or warnings,
we can now install our package with install(). Then, we can
attach the package with library() and use it. In RStudio
the Build menu has also functionality to check and install the
package.
install()
library(mypackage)
fnAreaCircle(2)After installing and attaching the package with
library(mypackage) we try to execute
fnAreaCircle() but we get an error.
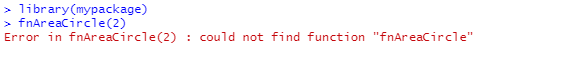
This is because by default, the functions in a package are only
available for internal use. We can make the functions available so users
can call them by putting @export in a roxygen comment above
the function. roxygen comments start with #' to distinguish
them from regular comments.
#' Calculate the area of a circle given its radius
#'
#' @export
fnAreaCircle <- function(r){
area <- pi*r^2
return(area)
}Then we run devtools::document() to (re)generate a
NAMESPACE file. We can learn more about roxygen and
NAMESPACE in the sections below.
devtools::document()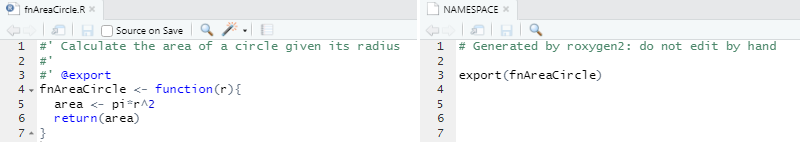
Then we check and re-install the package. We see that now the package works!
check()
install()
library(mypackage)
fnAreaCircle(2)
Exercise Find the folder where the new package has
been installed and inspect its components. .libPaths() can
be used to get the directory of the R packages.
Exercise Create a package mypackage2
that contains a function fnAreaRectangle() to calculate the
area of a rectangle given the length of its sides.
Just created my first R package!
meme package: https://cran.r-project.org/web/packages/meme/vignettes/meme.html
library(meme)
u <- system.file("success.jpg", package = "meme")
meme(u, "JUST CREATED MY FIRST", "R PACKAGE!")
3 R directory
The R/ subdirectory contains all the R code as a single
or multiple files.
Style
The tidyverse style guide: https://style.tidyverse.org/index.html
Hadley Wickham: http://adv-r.had.co.nz/Style.html
Coding club style: https://ourcodingclub.github.io/tutorials/etiquette/
styler: https://styler.r-lib.org/
Conditions
Errors, warnings and messages: https://adv-r.hadley.nz/conditions.html
4 DESCRIPTION
https://r-pkgs.org/description.html
The DESCRIPTION file stores metadata of our package such
as name, version number, author, license and dependencies on other
packages. The image below shows the DESCRIPTION file
created when executing create_package().
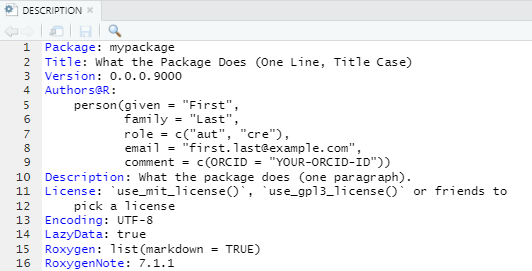
The DESCRIPTION file contains the fields
Imports and Suggests to declare the packages
that our package needs to work. Imports lists packages that
are strictly needed. Suggest lists packages that are not
strictly necessary but that can be needed to run tests or examples in
the documentation.
Packages listed in Imports are installed any time our
package is installed (but they are not be attached as when we execute
library()). Packages listed in Suggests are
not automatically installed when our package is installed.
DESCRIPTION can also include packages in
Depends to indicate dependency on a particular version of
R, and on packages that need to be attached with library()
when we use our package. More information about packages dependencies
can be read here and here.
When writing our package, the best practice is to refer to external
functions using package::function() to make it easy to
identify which functions are from other packages.
Packages in Imports and Suggests are
written as comma-separated list of package names. It is recommended
putting each package in one line and in alphabetical order. We can add
packages to Imports and Suggests with
usethis::use_package(). We can also execute
usethis::use_tidy_description() regularly to order and
format the DESCRIPTION fields. For example,
usethis::use_package("sf") # Default is "Imports"
usethis::use_package("leaflet")
usethis::use_package("ggplot2")
usethis::use_package("DT", "Suggests")
usethis::use_tidy_description()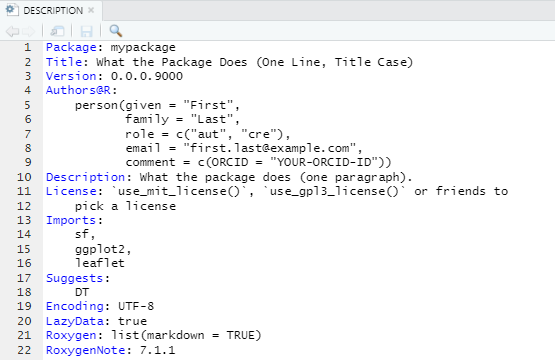
Exercise
Add packages to the Imports and Suggest
fields of the DESCRIPTION file.
5 NAMESPACE
https://r-pkgs.org/namespace.html
The NAMESPACE file specifies the functions in the
package that are exported to the user, and functions or packages that
are imported by the package. Exported functions are functions from our
package that are accessible by the user, and imported functions are
functions from other packages used by our package.
Below is the NAMESPACE file for the
mvtsplot package.
export("mvtsplot")
import(splines)
import(RColorBrewer)
importFrom("grDevices", "colorRampPalette", "gray")
importFrom("graphics", "abline", "axis", "box", "image", "layout",
"lines", "par", "plot", "points", "segments", "strwidth",
"text", "Axis")
importFrom("stats", "complete.cases", "lm", "na.exclude", "predict",
"quantile")Functions are exported to make it accessible by the user with
export(functioname).
import(packagename) allows us to access all exported
functions of an external package.
importFrom(packagename, functionames) imports the functions
of a specific package.
Every package mentioned in NAMESPACE must also be
present in the Imports or Depends fields of
DESCRIPTION.
:: operator
The :: operator can be used to access a function without
attaching the package. If the package is already installed, we could
call packagename::functionname(). Alternatively, we could
also execute library(packagename) first and then
functionname().
It is recommended to list packages in the Imports field
in DESCRIPTION so it is installed, but not in
NAMESPACE. Then we refer to the functions in the packages
explicitly with packagename::functioname().
The use of packagename::functioname() avoids confusion
in case there is more than one package with the same function name, and
clarifies the package the function belongs to.
@export
To export an object, we need to put @export in its
roxygen block and this will be added to NAMESPACE when
calling document().
6 Exercise
Create package mypackage with functions
fnAreaCircle() and fnAreaRectangle() to
calculate the area of a circle and rectangle, respectively.
library(devtools)
usethis::create_package("C://mypackage")
use_r("fnAreaCircle")
use_r("fnAreaRectangle")
document()
load_all()
fnAreaCircle(2)
fnAreaRectangle(2, 3)
use_mit_license()
check()
install()
library(mypackage)
fnAreaCircle(2)
#' Calculate the area of a circle given its radius
#'
#' @param r radius
#' @export
fnAreaCircle <- function(r){
area <- pi*r^2
return(area)
}
#' Calculate the area of a rectangle given its sides
#'
#' @param s1 side 1
#' @param s2 side 2
#' @export
fnAreaRectangle <- function(s1, s2){
area <- s1*s2
return(area)
}Add function fnBarplotAreas() to create a barplot with
ggplot2 with the areas of the circle and rectangle
use_r("fnBarplotAreas")
document()
#' Create ggplot with the areas of circle and rectangle
#'
#' @param acircle area circle
#' @param arectangle area rectangle
#' @export
fnBarplotAreas <- function(acircle, arectangle){
data <- data.frame(name = c("circle", "rectangle"), value = c(acircle, arectangle))
ggplot(data, aes(x = name, y = value)) + geom_bar(stat = "identity")
}When we execute the function we get an error because the function
uses ggplot2 and this package has not been attached.
load_all()
fnBarplotAreas(2,3)
# Error in ggplot(data, aes(x = name, y = value)) : could not find function "ggplot"To solve this error, in Imports in
DESCRIPTION, we add ggplot2 with
usethis::use_package("ggplot2"), and in
NAMESPACE we add import(ggplot2).
Note that if we only add ggplot2 in Imports
in DESCRIPTION but not import(ggplot2) in
NAMESPACE, ggplot2 will be installed but not
attached (with library()). Then, to use
fnBarplotAreas() we would need do
library(ggplot2) first, or we would need to call the
ggplot2 functions in fnBarPlotAreas() with the
:: operator.
7 Data
We can include data in our package in three ways:
- External data in
data/. These data are available to the user and can be, for instance, data for examples. Each file indata/should be a.rdafile created bysave()containing a single object with the same name as the file. - Internal data in
R/systdata.rda. These data are not available to the user and can be, for instance, data that the functions of our package need. - Raw data in
inst/extdata. When a package is installed, all files and folders ininst/are moved to the top-level directory, so raw data cannot have names likeR/orDESCRIPTION.
Data can be created with the following code:
# External data
x <- sample(1000)
usethis::use_data(x, mtcars)# Internal data
x <- sample(1000)
usethis::use_data(x, mtcars, internal = TRUE)We can refer to files in inst/extdata with
system.file(). For example:
system.file("extdata", "iris.csv", package = "readr", mustWork = TRUE)
# mustWork logical. If TRUE, an error is given if there are no matching files.The DESCRIPTION file can include
LazyData: true which means the datasets will be lazily
loaded, that is, they will not occupy any memory until we use them. When
we use usethis::create_package(),
LazyData: true will be included.
Exercise Inspect the data in the
ggplot2 package here https://github.com/tidyverse/ggplot2
Exercise Check the location of image files in the
meme package in GitHub (angry8.jpg, success.jpg, etc.) (https://github.com/GuangchuangYu/meme/tree/master/inst).
Then, check the location of image files in the meme package
in the installed package in your computer (meme/).
.libPaths() can be used to get the directory of the R
packages. We see that in GitHub images are in inst/ and in
the installed package images are in the top-level directory.
8 Documentation
The man/ directory contains the documentation files for
the exported objects of a package. To document the functions and data in
a package, we need .Rd files written in Latex-style
notation in the man/ directory. Instead of writing the
.Rd files by hand, we can add roxygen2 comments to the
.R files and then run devtools::document() to
convert the roxygen comments to .Rd files in
man/. roxygen comments start with #' to
distinguish them from regular comments and allow us to inclue tags
(e.g., @param, @return, etc.)
8.1 Documenting R functions
This is an example of documentation of a function:
#' Add together two numbers
#'
#' @param x A number.
#' @param y A number.
#' @return The sum of \code{x} and \code{y}.
#' @examples
#' add(1, 1)
#' add(10, 1)
add <- function(x, y) {
x + y
}To preview function documentation, we use ?fn or
help("fn"). This makes R to look for an .Rd
file in man/ containing \alias{"fn"}. Then
converts it into HTML and displays it.
8.2 Documenting a package
We can use roxygen to document a package as a whole and provide a
help page for our package. Package documentation can be accessed with
?packagename.
There is no object that corresponds to a package, so we need to
document NULL. We need to write
@docType package and @name packagename
Documentation of a package can be saved in a file called
packagename.R For example:
#' foo: A package for computating the notorious bar statistic
#'
#' The foo package provides three categories of important functions:
#' foo, bar and baz.
#'
#' @section Foo functions:
#' The foo functions ...
#'
#' @docType package
#' @name foo
NULL8.3 Documenting data
Objects in data/ are always exported and they must be
documented. Instead of documenting the data directly, we document the
name of the dataset and save it in R/. We never
@export a data set.
Below is an example of the documentation of the data
diamonds in ggplot2 which is saved as
R/data.R. More examples of the documentation of the data in
ggplot2 are here: https://github.com/tidyverse/ggplot2/blob/main/R/data.R
#' Prices of 50,000 round cut diamonds.
#'
#' A dataset containing the prices and other attributes of almost 54,000
#' diamonds.
#'
#' @format A data frame with 53940 rows and 10 variables:
#' \describe{
#' \item{price}{price, in US dollars}
#' \item{carat}{weight of the diamond, in carats}
#' ...
#' }
#' @source \url{http://www.diamondse.info/}
"diamonds"9 Vignettes
Our package can also include vignettes that demonstrate how to use
the functions of the package to solve a specific problem. We can see the
vignettes of a package by typing
browseVignettes("packagename").
We can write a vignette using R Markdown and knitr. We
can start creating a vignette by running
usethis::use_vignette("myvignette")This creates a vignettes/ directory and drafts the
vignette vignettes/myvignette.Rmd. It also adds the
necessary dependencies to DESCRIPTION (adds knitr to the
Suggests and VignetteBuilder fields).
The first few lines of the vignette will be a YAML header with metadata as follows:
---
title: "Vignette Title"
output: rmarkdown::html_vignette
vignette: >
%\VignetteIndexEntry{Vignette Title}
%\VignetteEngine{knitr::rmarkdown}
\usepackage[utf8]{inputenc}
---Here, symbol > indicates the following lines are
plain text that do not use any special YAML features. After the header
we write the vignette using R Markdown intermingling text and R
code.
We can run the vignette by using knitr. We can use
devtools::build_vignettes() to build vignettes. The
resulting .HTML vignette will be created in the
inst/doc folder. Alternatively, by executing
devtools::build() the .HTML vignette will be
built as part of the construction of the .tar.gz file for
the package bundle.
10 Testing
It is important to test the R code in the package to make sure there are no bugs and everything works as expected.
When we are developing a package, we test the code in an informal way. It is worth it to keep these tests as part of our package so we can rerun them whenever is needed to ensure the package continues working well and discover bugs in the code.
The package testthat allows us to write automated tests
about expectations on how the functions of our package should work. That
is, we can check that given a set of inputs, the functions return the
expected outputs.
We can set up our package to use testthat by
running:
usethis::use_testthat()This creates a tests/testhat directory, adds
testthat to the Suggest field in the
DESCRIPTION, and creates a file
tests/testthat.R that runs all tests when we check our
package.
Then we need to write our tests in test/testhtat and run
them with devtools::test().
Tests are organized in a hierarchical way, namely, expectations are grouped in tests, and tests are grouped in files.
Expectations are functions that describe the expected result of a computation. Expectations start with
expect_and have two arguments: the actual result and what we expect. If the actual and expected result do not agreetestthatthrows an error.A test groups together a set of expectations to test a unit of functionality. A test is created with
test_that()passing the test name and code block as arguments.Then tests are organized in files which are given a name with
context().
Below is an example of a test file from the stringr
package:
context("String length")
library(stringr)
test_that("str_length is number of characters", {
expect_equal(str_length("a"), 1)
expect_equal(str_length("ab"), 2)
expect_equal(str_length("abc"), 3)
})
#> Test passed 😸
test_that("str_length of factor is length of level", {
expect_equal(str_length(factor("a")), 1)
expect_equal(str_length(factor("ab")), 2)
expect_equal(str_length(factor("abc")), 3)
})
#> Test passed 🥇
test_that("str_length of missing is missing", {
expect_equal(str_length(NA), NA_integer_)
expect_equal(str_length(c(NA, 1)), c(NA, 1))
expect_equal(str_length("NA"), 2)
})
#> Test passed 🎉11 License
https://r-pkgs.org/license.html
License for the R package is specified in the
DESCRIPTION file in License: typelicense. We
can also specify License: file LICENSE, and create a text
file called LICENSE that explains the type of license.
The license contains how the code can be inspected, modified, and
distributed. The usethis package has several functions to
create a number of licenses including the following:
MIT license (
use_mit_license()) is a permissive license that allows people to use the code with minimal restrictionsGPLv3 license (
use_gpl_license()) is a copyleft license so that all derivatives of the code are also open sourceIf we do not want to make the code open source we can use
use_proprietary_license(). These packages cannot be distributed by CRANFor packages that primarily contain data, not code, we can choose Creative Commons licenses such as the CC0 license (
use_cc0_license()) if we want minimal restrictions, or the CC BY license (use_ccby_license()) if we require attribution when someones uses the data
12 Citing packages
https://r-pkgs.org/inst.html?q=citation#inst-citation
In R, we can execute citation() to get a citation of R,
and citation("packagename") to get a citation of a
particular package.
We can add a citation for our package by adding a file in
inst/CITATION similar to this:
citHeader("To cite lubridate in publications use:")
citEntry(entry = "Article",
title = "Dates and Times Made Easy with {lubridate}",
author = personList(as.person("Garrett Grolemund"),
as.person("Hadley Wickham")),
journal = "Journal of Statistical Software",
year = "2011",
volume = "40",
number = "3",
pages = "1--25",
url = "https://www.jstatsoft.org/v40/i03/",
textVersion =
paste("Garrett Grolemund, Hadley Wickham (2011).",
"Dates and Times Made Easy with lubridate.",
"Journal of Statistical Software, 40(3), 1-25.",
"URL https://www.jstatsoft.org/v40/i03/.")
)13 README.Rmd
We can create a README.Rmd file with a description of
the package, installation instructions and basic examples.
usethis::use_readme_rmd() creates a template
README.Rmd and adds it to .Rbuildignore.
devtools::build_readme() can be used to render the document
to README.md. README.md is rendered by GitHub
on the main page.
14 NEWS.md
NEWS.md can be created to track the changes from one
version of a package to another. We can create this file with
usethis::use_news_md().
15 Naming a package
https://r-pkgs.org/workflows101.html
A package name can only consist of letters, numbers and periods (no
- or _). The name must start with a letter and
cannot end with a period. The package available function
can be used to see whether a package name is available.
library(available)
available("packagename")17 Submission to CRAN
CRAN is the main repository for R packages. When R packages are contributed to CRAN, R users can easily discover and install them with
install.packages("packagename")You can read about the submission process to CRAN here: https://r-pkgs.org/release.html
18 Software review and best practices
rOpenSci helps develop R packages for the sciences via community driven learning, review and maintenance of contributed software in the R ecosystem.
- rOpenSci: https://ropensci.org/
- rOpenSci Packages: Development, Maintenance, and Peer Review: https://devguide.ropensci.org/
- rOpenSci Statistical Software Peer Review: https://stats-devguide.ropensci.org/
Submission of a package
Guide for authors: https://devguide.ropensci.org/authors-guide.html
Open a new issue in the software review repository and fill out the template: https://github.com/ropensci/software-review/issues/new/choose
Example: https://github.com/ropensci/software-review/issues/160#issuecomment-355043656
Issues: https://github.com/ropensci/software-review/issues
rOpenSci packages: https://ropensci.org/packages/all/
19 Websites for packages
with the pkgdown package
- Package websites with pkgdown: https://pkgdown.r-lib.org/index.html
- Introduction to pkgdown: https://pkgdown.r-lib.org/articles/pkgdown.html
To create a website for our package with pkgdown, we
need to have an R package with a local directory and a GitHub
repository. Then, from within the package directory we run:
# Install released version from CRAN
install.packages("pkgdown")
# Run once to configure your package to use pkgdown
usethis::use_pkgdown()
# Build website
pkgdown::build_site()This generates a docs/ directory with the website. The
README.md is shown in the homepage, the documentation in
man/ generates a function reference, and the vignettes are
shown into articles/.
We can customize the website following https://pkgdown.r-lib.org/articles/pkgdown.html
We can publish the website online in GitHub Pages. When pushing the
changes, we need to make sure the docs directory does not
appear in the .gitignore file. Then we need to configure
GitHub Pages to use the docs directory.
Settings > Pages Branch: master /docs
Your site is ready to be published at http://www.paulamoraga.com/mypackage/
20 Hexstickers
hexstickerfor packages: https://github.com/GuangchuangYu/hexSticker
21 Exercise
Create a repository in GitHub
In GitHub, create a repository with name mypackage
(do not add README and .gitignore files.
This will be added later)
Copy SSH git@github.com:Paula-Moraga/mypackage.git
Create project in RStudio
File > New Project > Version Control > Git. Copy repository URL.
Repository URL:
git@github.com:Paula-Moraga/mypackage.git
Project directory name: mypackage
Create package
library(devtools)
create_package(getwd())Question: Overwrite pre-existing file
mypackage.Rproj?
Select: No
A second session of RStudio opens with a Build tab for building packages. We close the other RStudio session.
library(devtools)
use_r("fnAreaCircle")
use_mit_license()
usethis::use_vignette("myvignette")
devtools::build_vignettes()
usethis::use_readme_rmd()
devtools::build_readme()
document()
load_all()
fnAreaCircle(2)
check()#' Calculate the area of a circle given its radius
#'
#' @param r radius
#' @return area circle
#' @export
fnAreaCircle <- function(r){
area <- pi*r^2
return(area)
}Push to GitHub
In Terminal
git status
git add .
git commit -m "initial commit"
git pushCreating website
# Run once to configure your package to use pkgdown
usethis::use_pkgdown()
# Build website
pkgdown::build_site()This generates a docs/ directory with the website.
Push to GitHub
When pushing the changes to GitHub, delete docs
directory from .gitignore.
Publish website in GitHub Pages
Configure GitHub Pages to use the docs directory.
Settings > Pages Branch: master /docs
Your site is ready to be published at http://www.paulamoraga.com/mypackage/
Install mypackage from GitHub
devtools::install_github("Paula-Moraga/mypackage")
library(mypackage)
?fnAreaCircle
fnAreaCircle(2)22 Installing and attaching R packages
To install an R package from CRAN, we need to use the
install.packages() function passing the name of the package
as first argument. Then, to use the package, we need to attach it using
the library() function. For example, to install and attach
the sf package, we need to type
install.packages("sf")
library(sf)A summary of useful functions to install and attach packages are the following:
install.packages("packagename"): install a package from
CRAN
devtools::install_github("username/packagename"):
install a package from GitHub
install.packages("packagenameversion.tar.gz", type = "source", repos = NULL):
install packages from source. This can be useful when we do not have
internet connection and have the source files locally
remove.packages("packagename"): remove an installed
package
update.packages(oldPkgs = "packagename"): update a
package
library("packagename"): attach a package (to start using
it)
detach("packagename"): dettach a package
installed.packages(): lists the installed packages. This
list can also be seen in the Packages tab of RStudio or by typing
library().
search(): lists the attached packages
sessionInfo(): version information about R, the OS and
attached or loaded packages
.libPaths(): path where R packages are installed. If we
want to change the default installation path, we can do it by editing
the .Rprofile file with
.libPaths("/pathinstallation/R").
usethis::edit_r_profile() opens .Rprofile.
library() and
require()
library() and require() both attach
packages.
require() tries to load the package using
library() and returns a logical value indicating the
success (TRUE) or failure (FALSE)
If we try to attach a package that has not been installed:
library()will throw and error and the program will stop.require()will throw a warning but not an error and will the program will continue running. The program will only stop when we try to use a function from the library that was not installed.
require() returns (invisibly) TRUE if the package is
available, and FALSE if the package is not available. We can install a
package if it does not exist and then attach it as follows:
if (!require(packagename)){
install.packages('package')
}
library(package)The following code allows us to install and attach a set of R packages in an efficient way:
# Package names
packages <- c("pkg1", "pkg2", "pkg3")
# Install packages not yet installed
installed_packages <- packages %in% rownames(installed.packages())
if (any(installed_packages == FALSE)) {
install.packages(packages[!installed_packages])
}
# Attach packages
invisible(lapply(packages, library, character.only = TRUE))
# invisible() removes the output
# character.only https://stackoverflow.com/questions/15273635/cant-figure-out-error-in-lapply